转录组数据—质量控制(数据质量评估,过滤低质量)
原创转录组数据—质量控制(数据质量评估,过滤低质量)
原创用户10412487
发布于 2023-04-19 21:27:49
发布于 2023-04-19 21:27:49
数据质量评估
软件Fastqc

fastqc 常用参数
(rna) Mar402 20:38:07 ~/project/Human-16-Asthma-Trans/data/rawdata #-t 6 同时对这6个文件进行质控 -o将文件的输出结果输出到...,注意要在数据所在目录下
$ fastqc -t 6 -o ./ SRR*.fastq.gz
application/gzip
application/gzip
Started analysis of SRR1039510_1.fastq.gz
application/gzip
application/gzip
application/gzip
application/gzip
Approx 5% complete for SRR1039510_1.fastq.gz
Approx 10% complete for SRR1039510_1.fastq.gz
Approx 15% complete for SRR1039510_1.fastq.gz
Approx 20% complete for SRR1039510_1.fastq.gz
Approx 25% complete for SRR1039510_1.fastq.gz
Approx 30% complete for SRR1039510_1.fastq.gz
Approx 35% complete for SRR1039510_1.fastq.gz
Approx 40% complete for SRR1039510_1.fastq.gz
Approx 45% complete for SRR1039510_1.fastq.gz
Approx 50% complete for SRR1039510_1.fastq.gz
Approx 55% complete for SRR1039510_1.fastq.gz
Approx 60% complete for SRR1039510_1.fastq.gz
Approx 65% complete for SRR1039510_1.fastq.gz
Approx 70% complete for SRR1039510_1.fastq.gz
Approx 75% complete for SRR1039510_1.fastq.gz
Approx 80% complete for SRR1039510_1.fastq.gz
Approx 85% complete for SRR1039510_1.fastq.gz
Approx 90% complete for SRR1039510_1.fastq.gz
Approx 95% complete for SRR1039510_1.fastq.gz
Approx 100% complete for SRR1039510_1.fastq.gz
Analysis complete for SRR1039510_1.fastq.gz
Started analysis of SRR1039510_2.fastq.gz
Approx 5% complete for SRR1039510_2.fastq.gz
Approx 10% complete for SRR1039510_2.fastq.gz
Approx 15% complete for SRR1039510_2.fastq.gz
Approx 20% complete for SRR1039510_2.fastq.gz
Approx 25% complete for SRR1039510_2.fastq.gz
Approx 30% complete for SRR1039510_2.fastq.gz
Approx 35% complete for SRR1039510_2.fastq.gz
Approx 40% complete for SRR1039510_2.fastq.gz
Approx 45% complete for SRR1039510_2.fastq.gz
Approx 50% complete for SRR1039510_2.fastq.gz
Approx 55% complete for SRR1039510_2.fastq.gz
Approx 60% complete for SRR1039510_2.fastq.gz
Approx 65% complete for SRR1039510_2.fastq.gz
Approx 70% complete for SRR1039510_2.fastq.gz
Approx 75% complete for SRR1039510_2.fastq.gz
Approx 80% complete for SRR1039510_2.fastq.gz
Approx 85% complete for SRR1039510_2.fastq.gz
Approx 90% complete for SRR1039510_2.fastq.gz
Approx 95% complete for SRR1039510_2.fastq.gz
Approx 100% complete for SRR1039510_2.fastq.gz
Analysis complete for SRR1039510_2.fastq.gz
Started analysis of SRR1039511_1.fastq.gz
Approx 5% complete for SRR1039511_1.fastq.gz
Approx 10% complete for SRR1039511_1.fastq.gz
Approx 15% complete for SRR1039511_1.fastq.gz
Approx 20% complete for SRR1039511_1.fastq.gz
Approx 25% complete for SRR1039511_1.fastq.gz
Approx 30% complete for SRR1039511_1.fastq.gz
Approx 35% complete for SRR1039511_1.fastq.gz
Approx 40% complete for SRR1039511_1.fastq.gz
Approx 45% complete for SRR1039511_1.fastq.gz
Approx 50% complete for SRR1039511_1.fastq.gz
Approx 55% complete for SRR1039511_1.fastq.gz
Approx 60% complete for SRR1039511_1.fastq.gz
Approx 65% complete for SRR1039511_1.fastq.gz
Approx 70% complete for SRR1039511_1.fastq.gz
Approx 75% complete for SRR1039511_1.fastq.gz
Approx 80% complete for SRR1039511_1.fastq.gz
Approx 85% complete for SRR1039511_1.fastq.gz
Approx 90% complete for SRR1039511_1.fastq.gz
Approx 95% complete for SRR1039511_1.fastq.gz
.......
#这是软件运行的动态日志
(rna) Mar402 20:38:17 ~/project/Human-16-Asthma-Trans/data/rawdata
$ ls #ls 后列出这些文件
SRR1039510_1_fastqc.html SRR1039510_2.fastq.gz SRR1039511_2_fastqc.zip SRR1039512_2_fastqc.html
SRR1039510_1_fastqc.zip SRR1039511_1_fastqc.html SRR1039511_2.fastq.gz SRR1039512_2_fastqc.zip
SRR1039510_1.fastq.gz SRR1039511_1_fastqc.zip SRR1039512_1_fastqc.html SRR1039512_2.fastq.gz
SRR1039510_2_fastqc.html SRR1039511_1.fastq.gz SRR1039512_1_fastqc.zip
SRR1039510_2_fastqc.zip SRR1039511_2_fastqc.html SRR1039512_1.fastq.gzfastqc运行
#方法一:直接运行 #缺点霸占控制台和时间
fastqc -t 6 -o ./ SRR*.fastq.gz
#方法二:在命令前后加上nohop & 使用FastQC软件对单个fastq文件进行质量评估,结果输出到qc/文件夹下(nohop no hang up 不挂起,退出终端不会影响程序的运行;&: 后台运行)(在后台运行!)(适用于比较长的简单的命令)
nohup fastqc -t 6 -o ./ SRR*.fastq.gz >qc.log &
#方法三:将命令写入shell脚本,使用nohop &运行sh脚本 使用MultiQc整合FastQC结果(适用于比较长的复杂的命令)
multiqc *.zip -o ./ 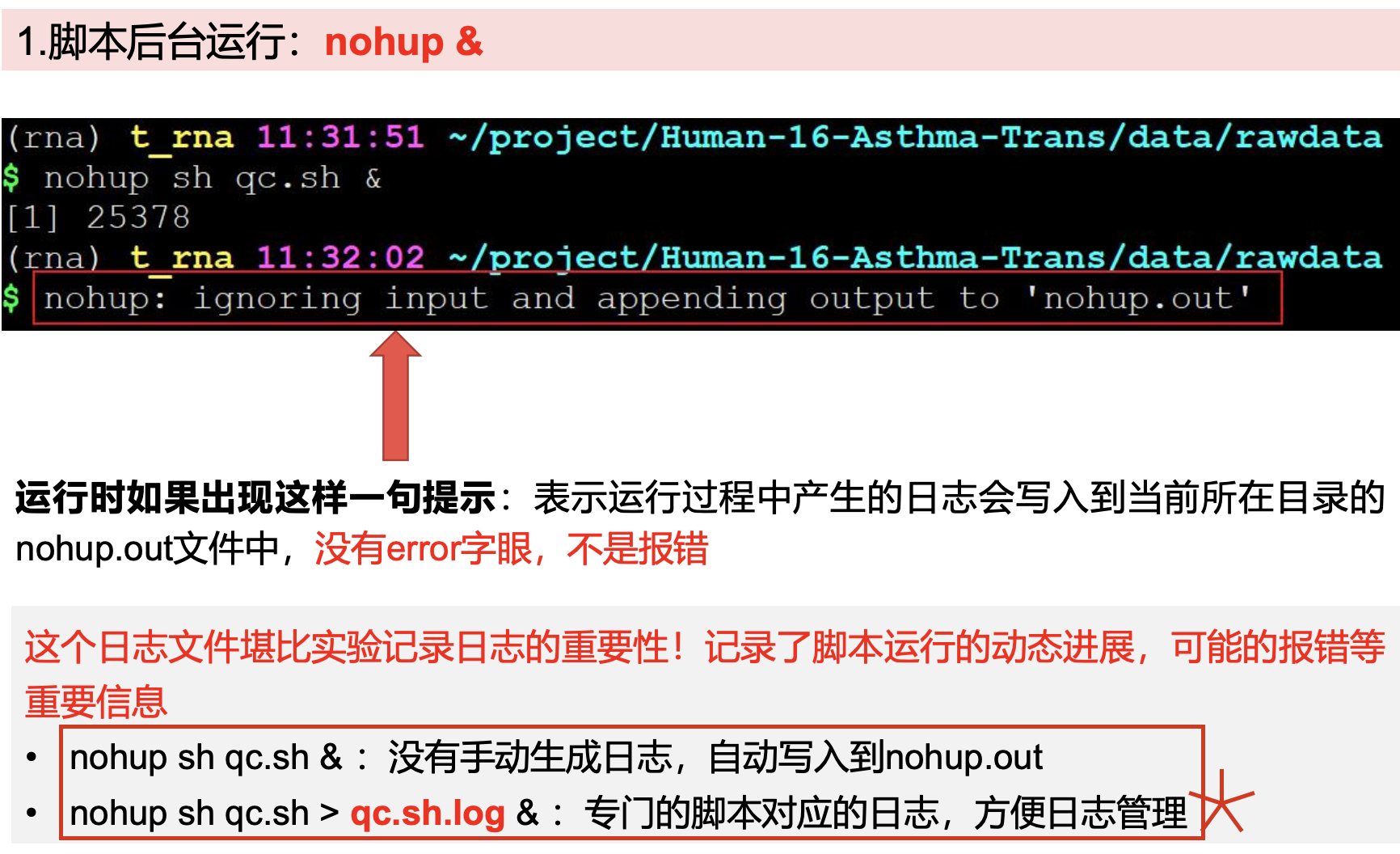
运行fastqc要写对应的日志
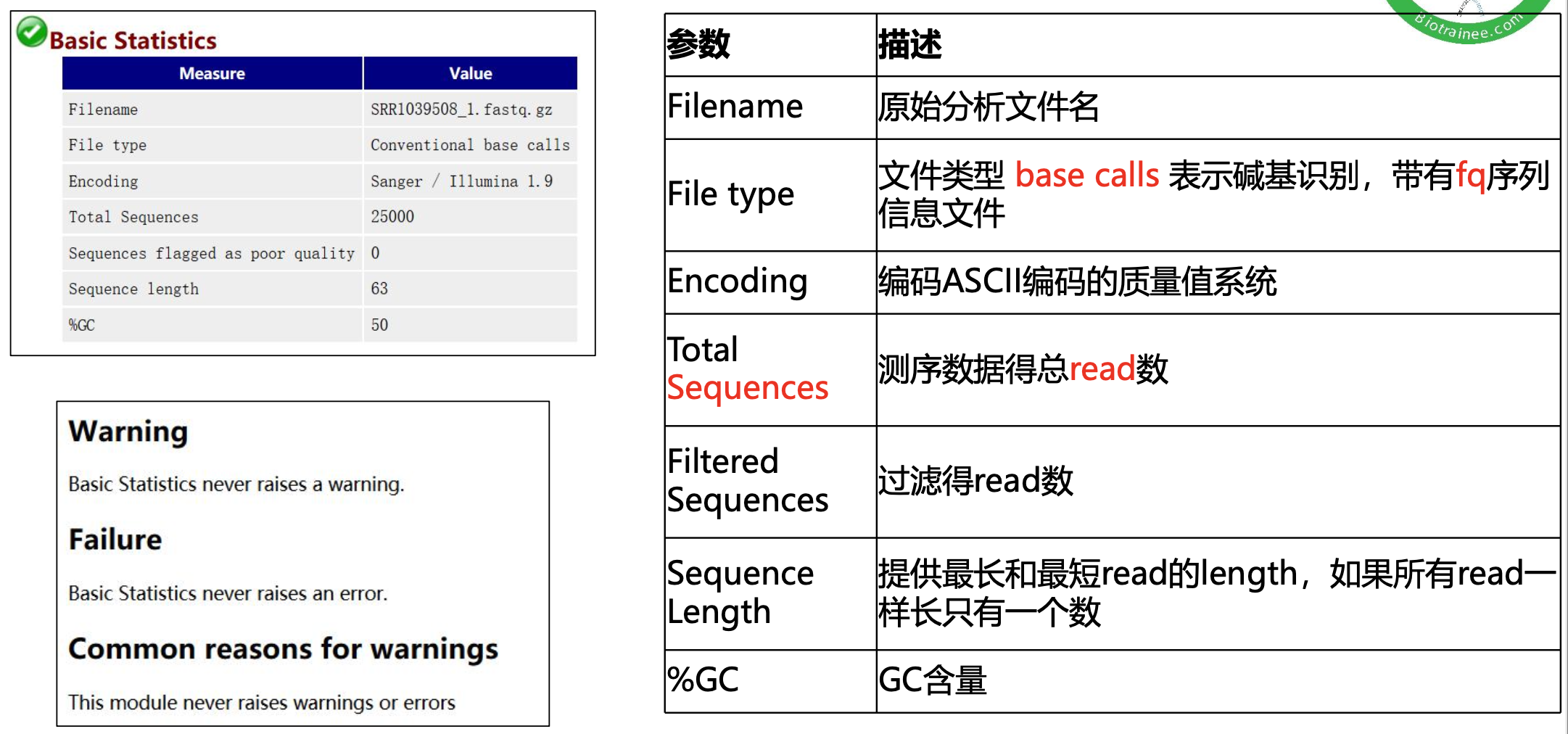
·fastqc后报告结果带有fastqc结尾的文件,html为主要质控报告,网页版本,使用浏览器打开;zip里面是表格或者图片等
·解压 *.fastqc.zip 得到pic1
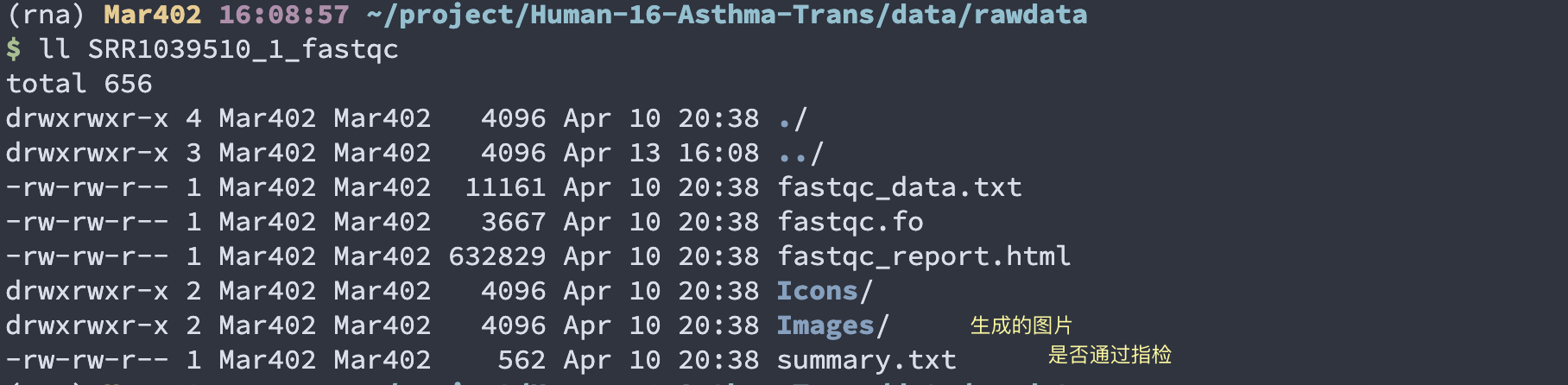
pic1
将质控报告下载至本地
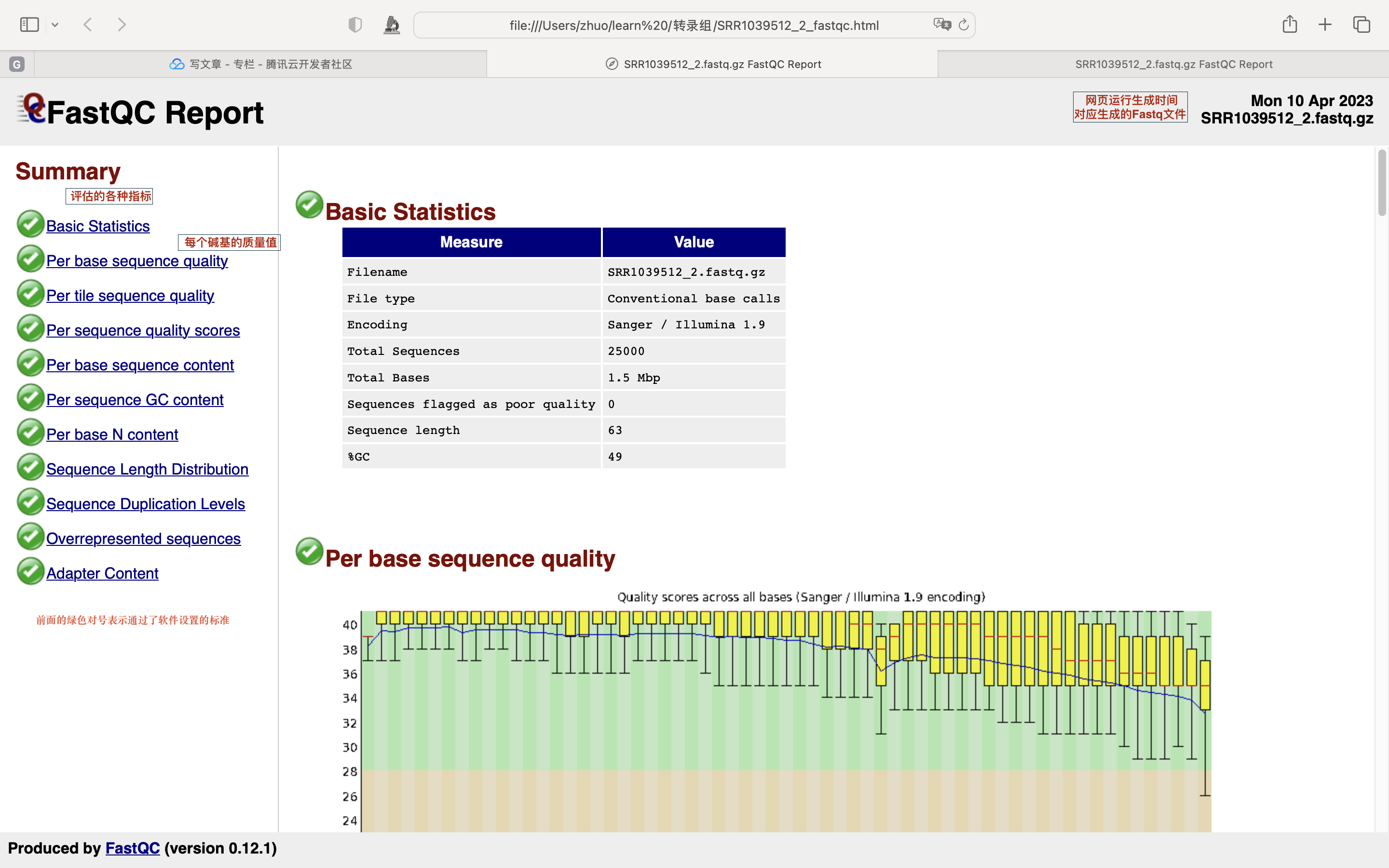
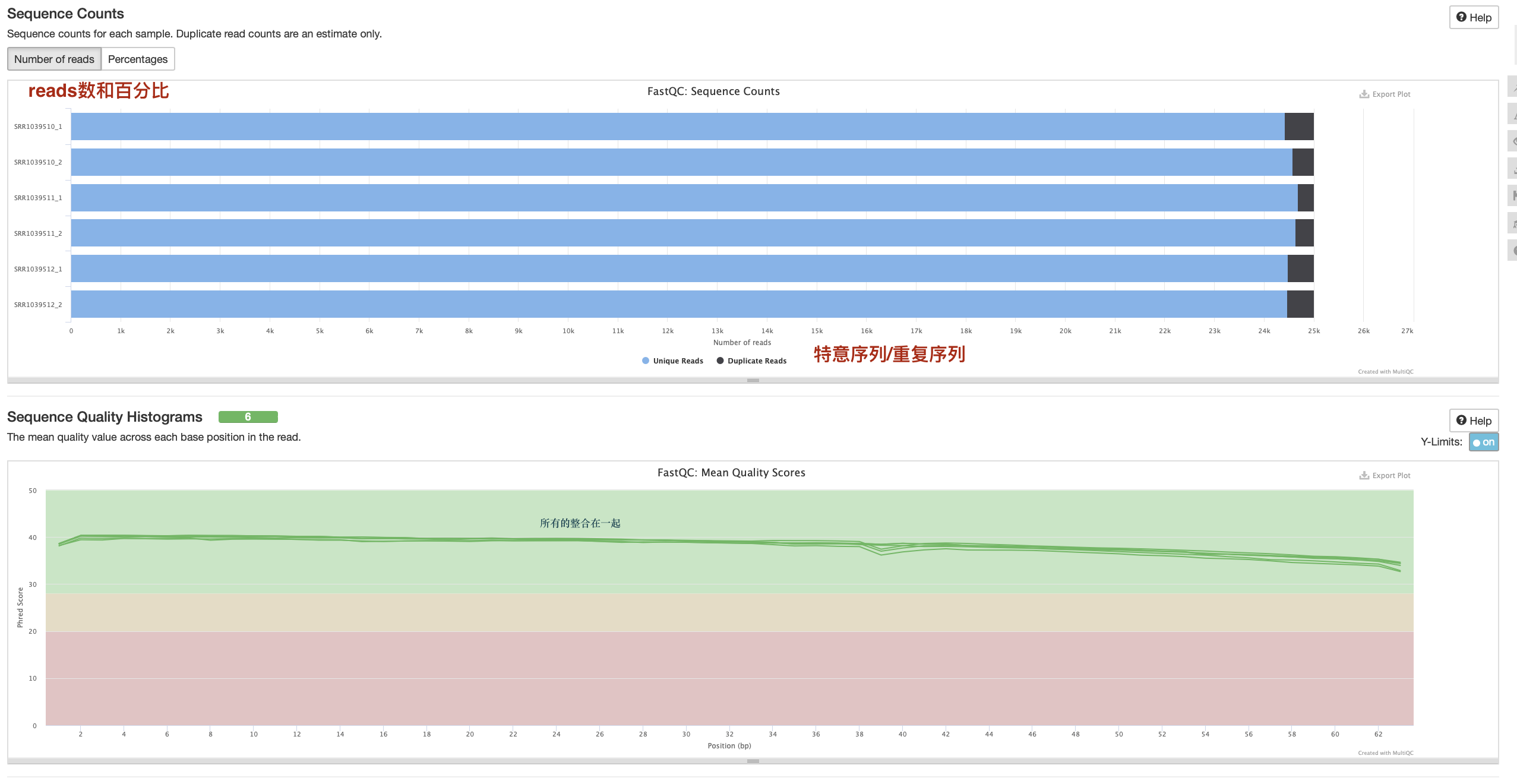
数据质控
数据量统计方式
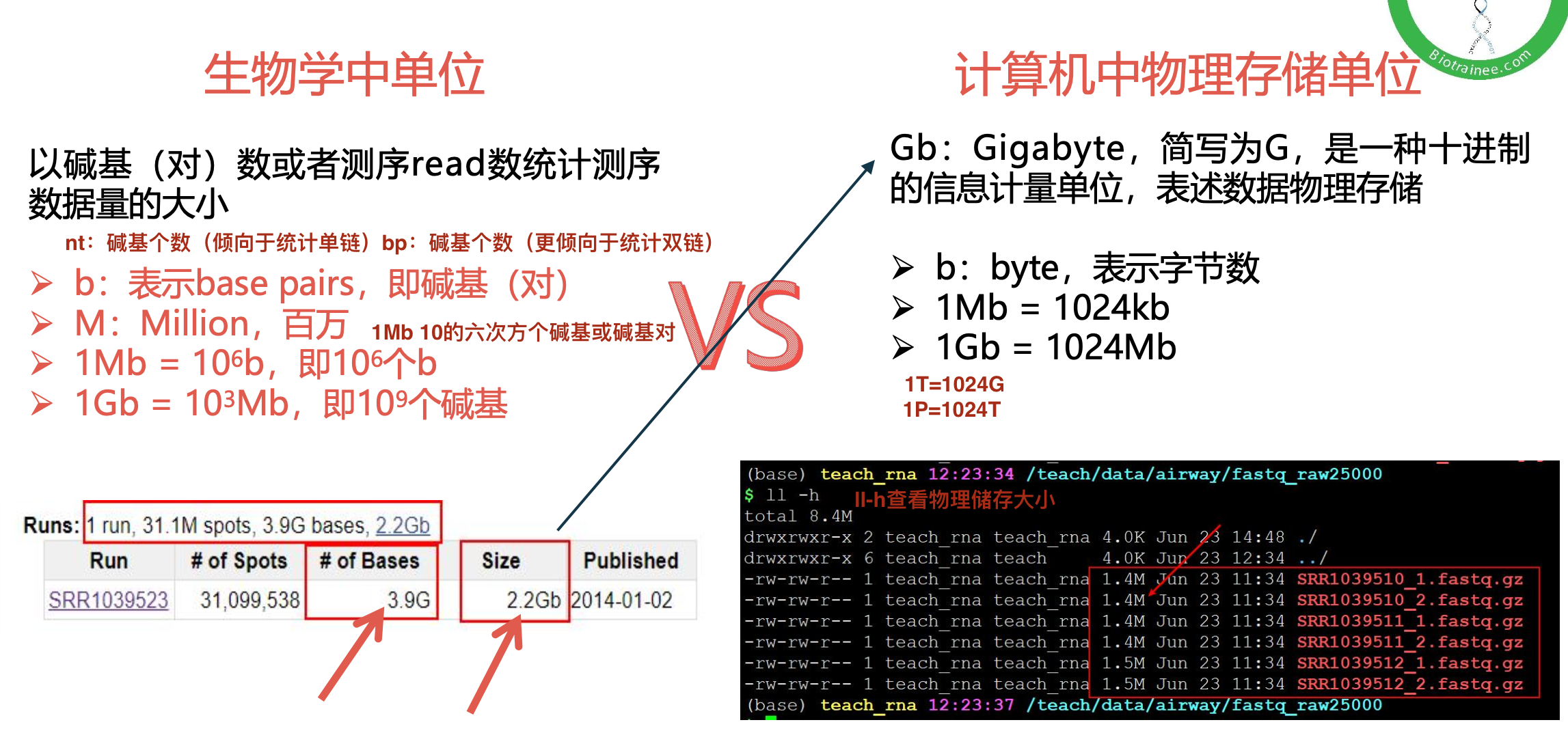
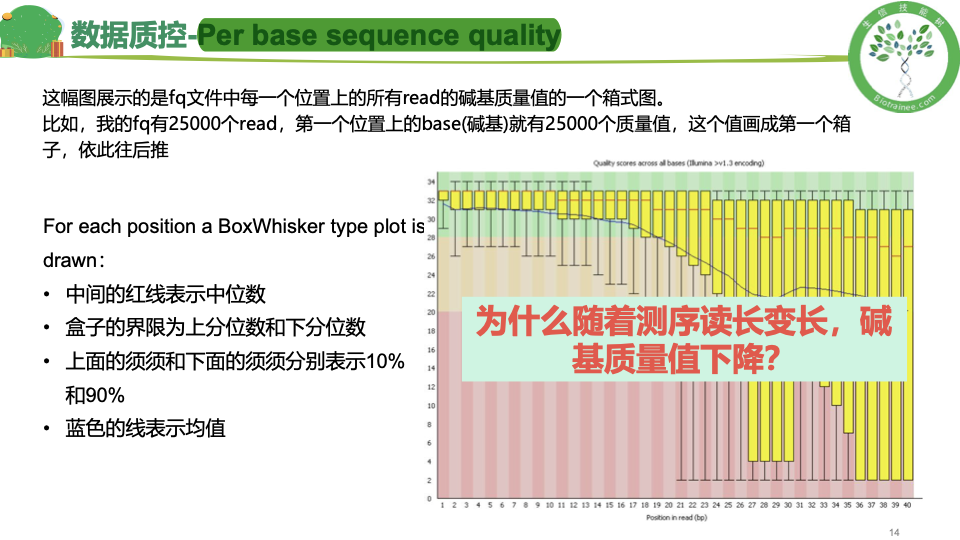
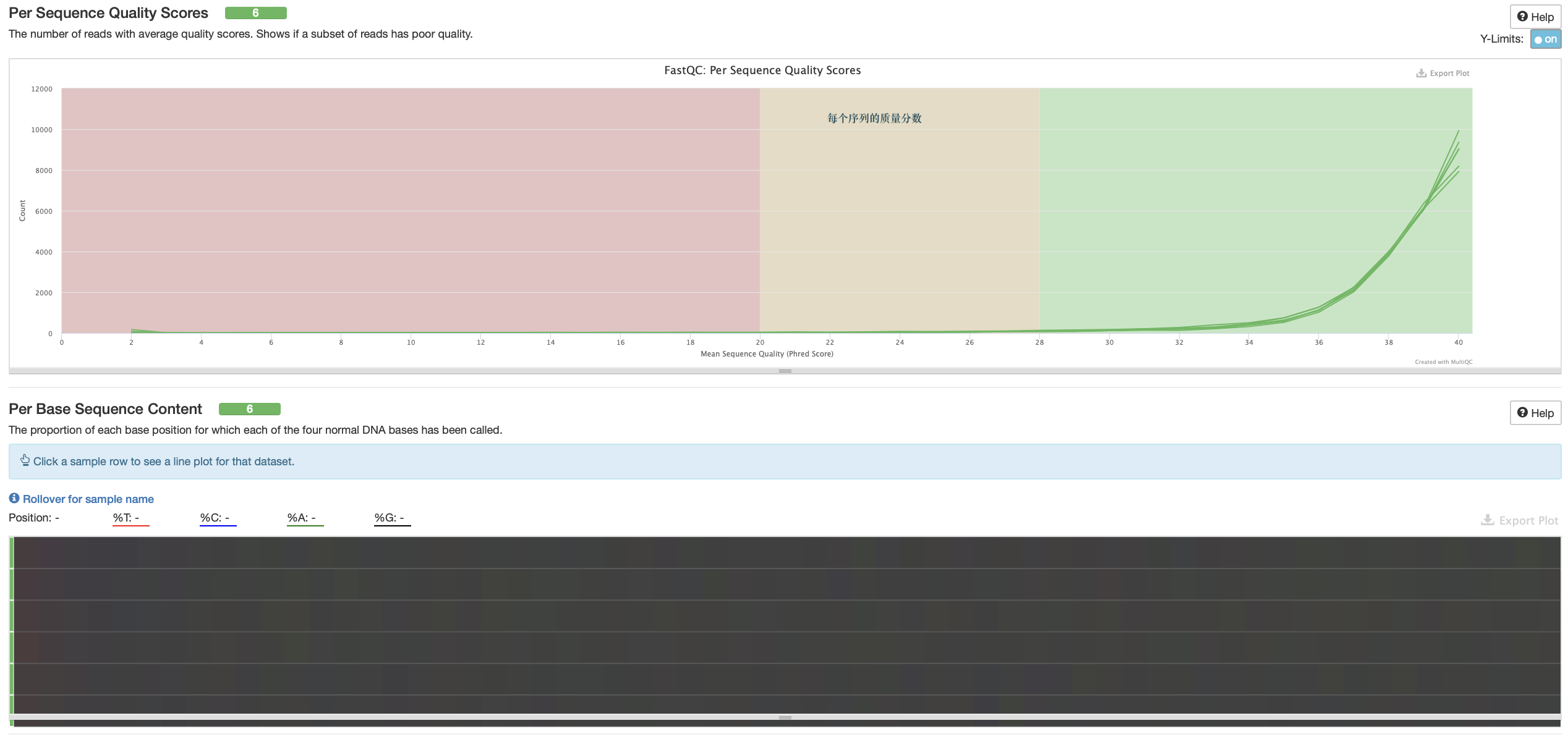
数据质控 PerPer base sequence quality

第二张图
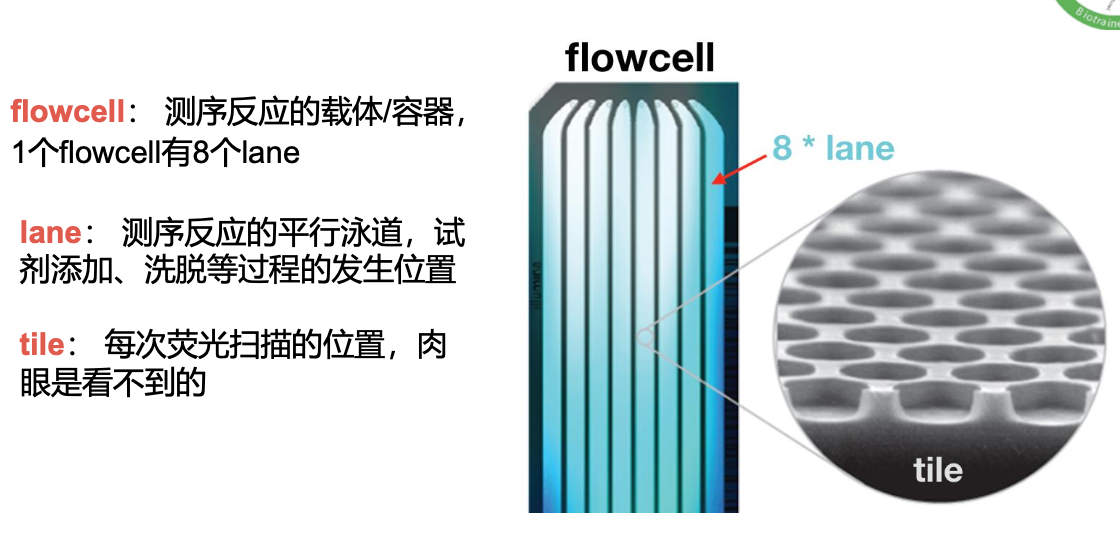
flowcell
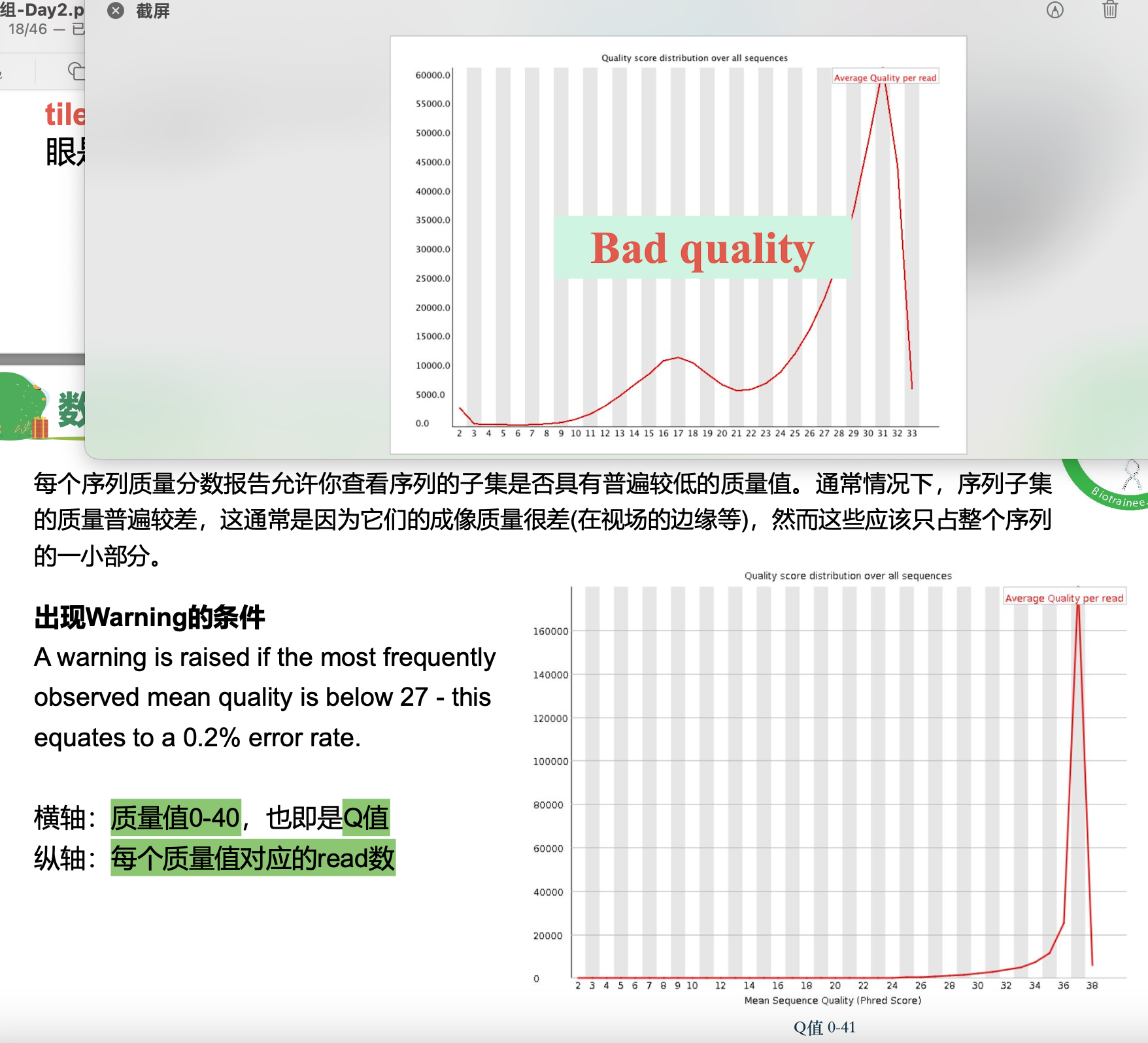
Q值图
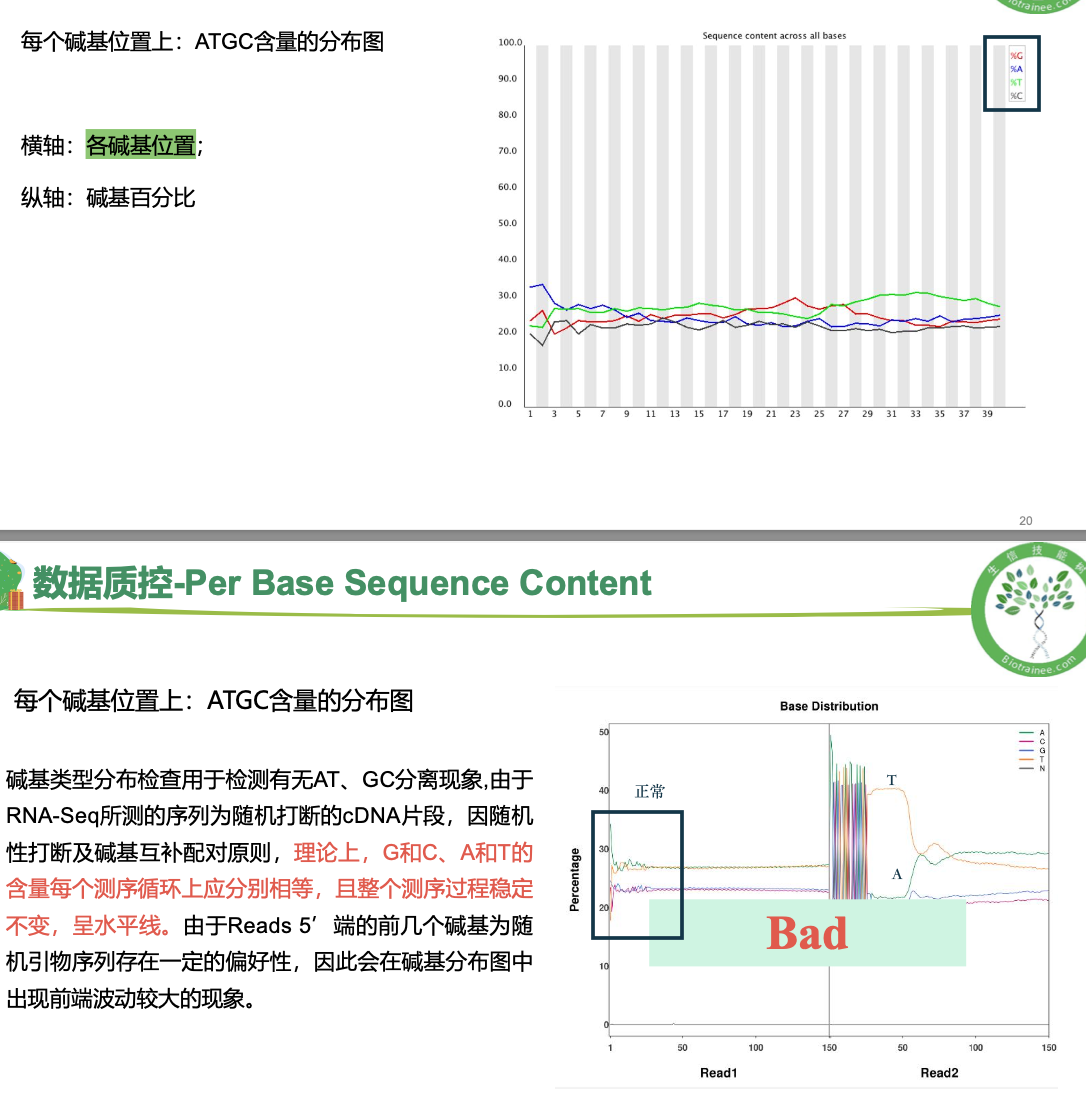
per base sequence
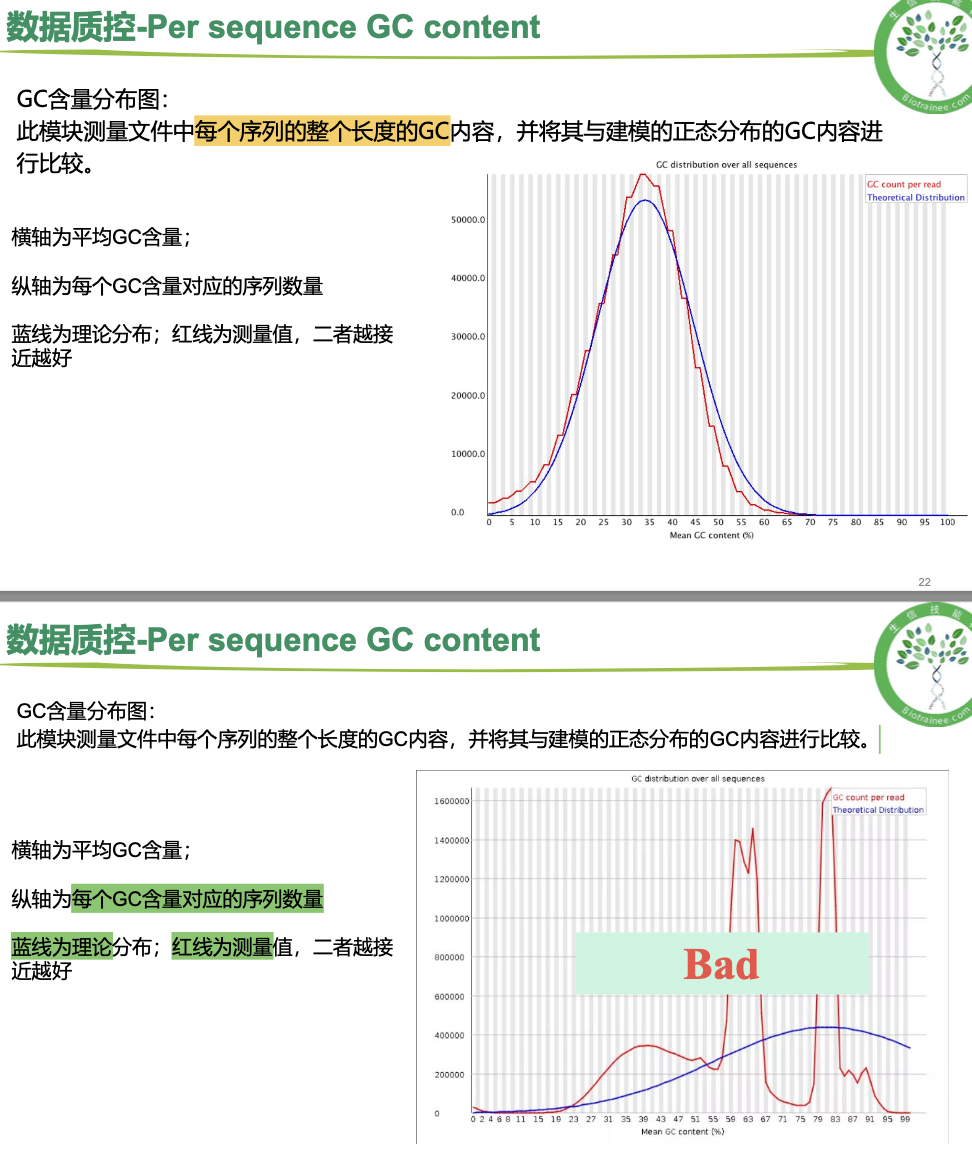
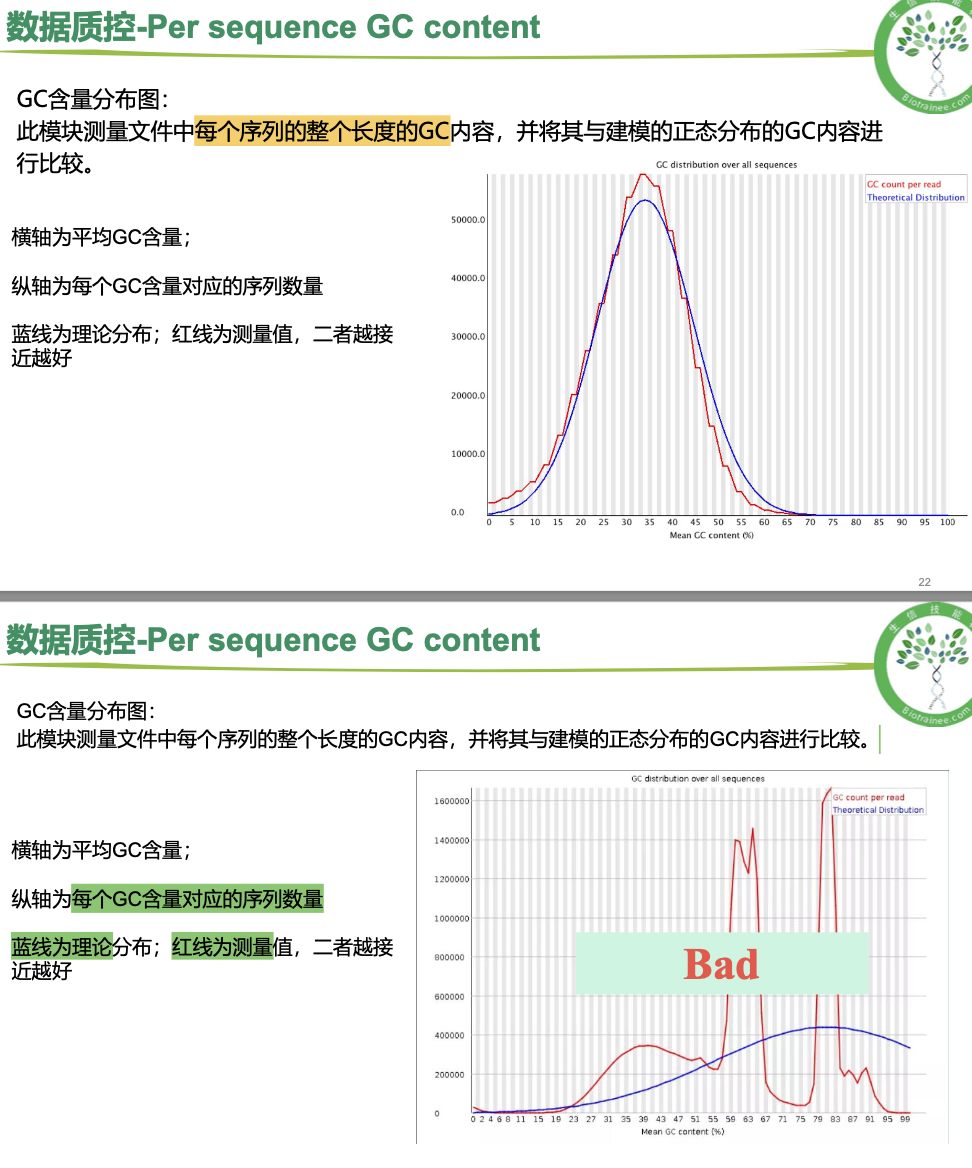
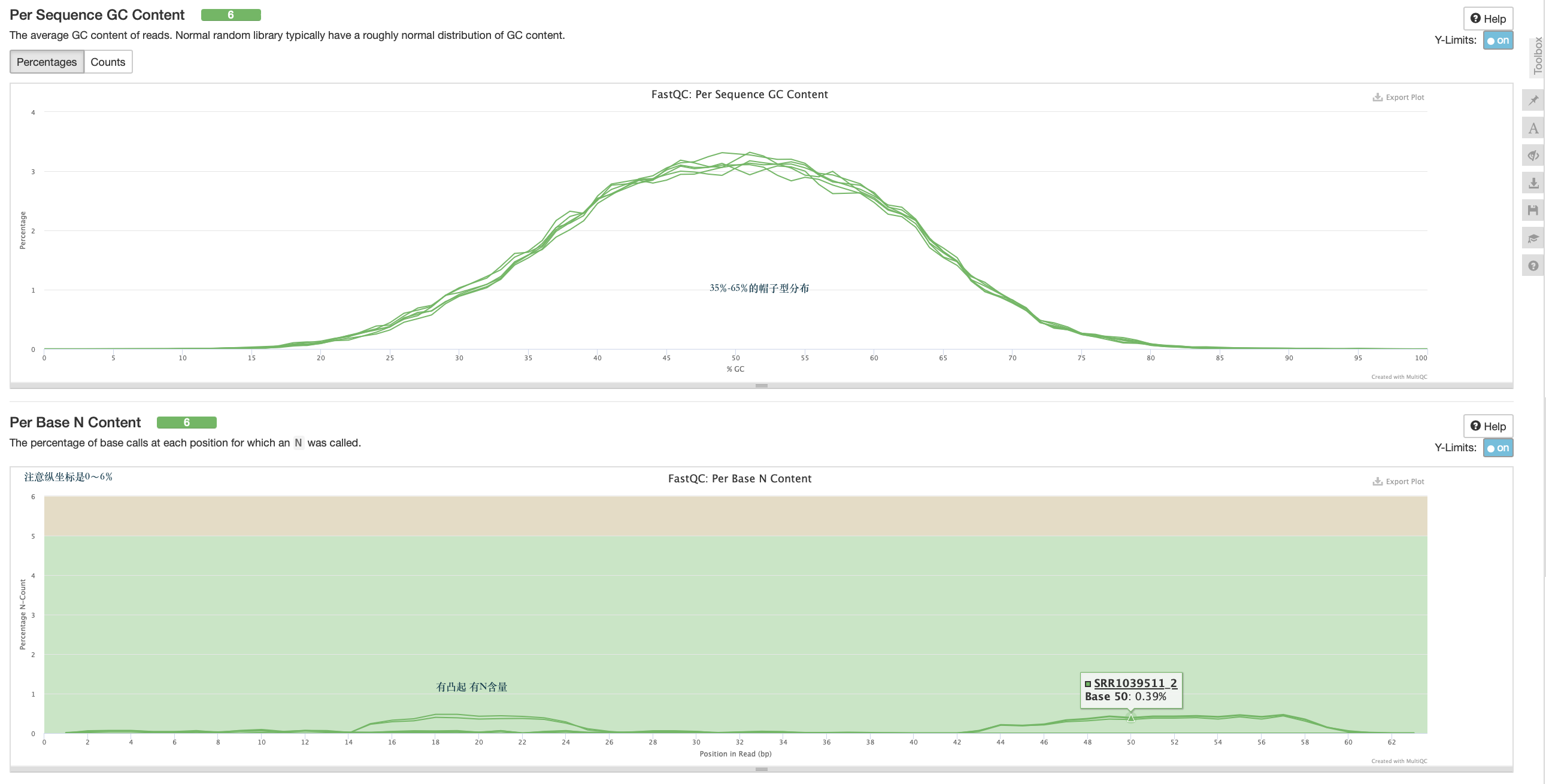
per sequence GC content
N 的含量
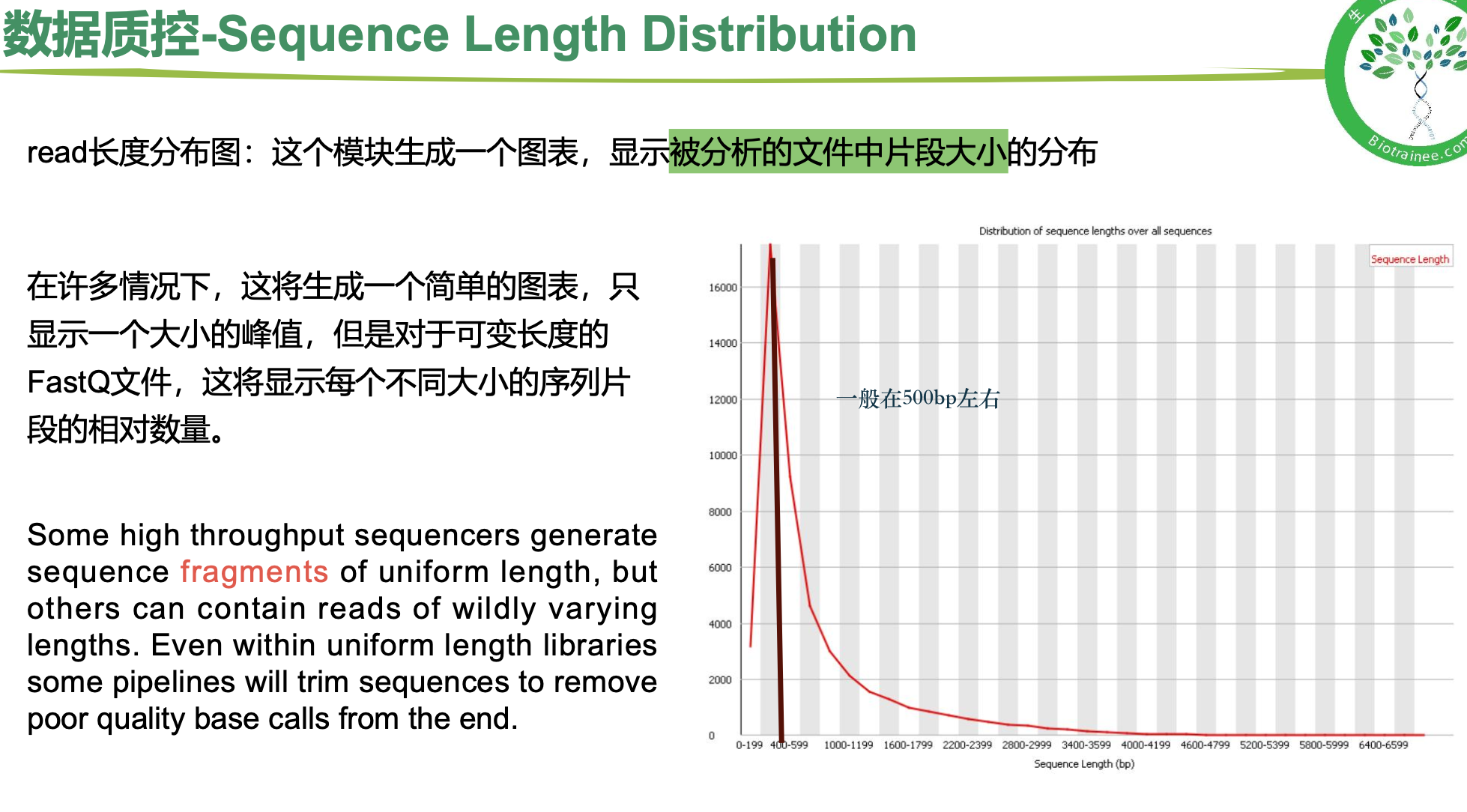
序列长度分布
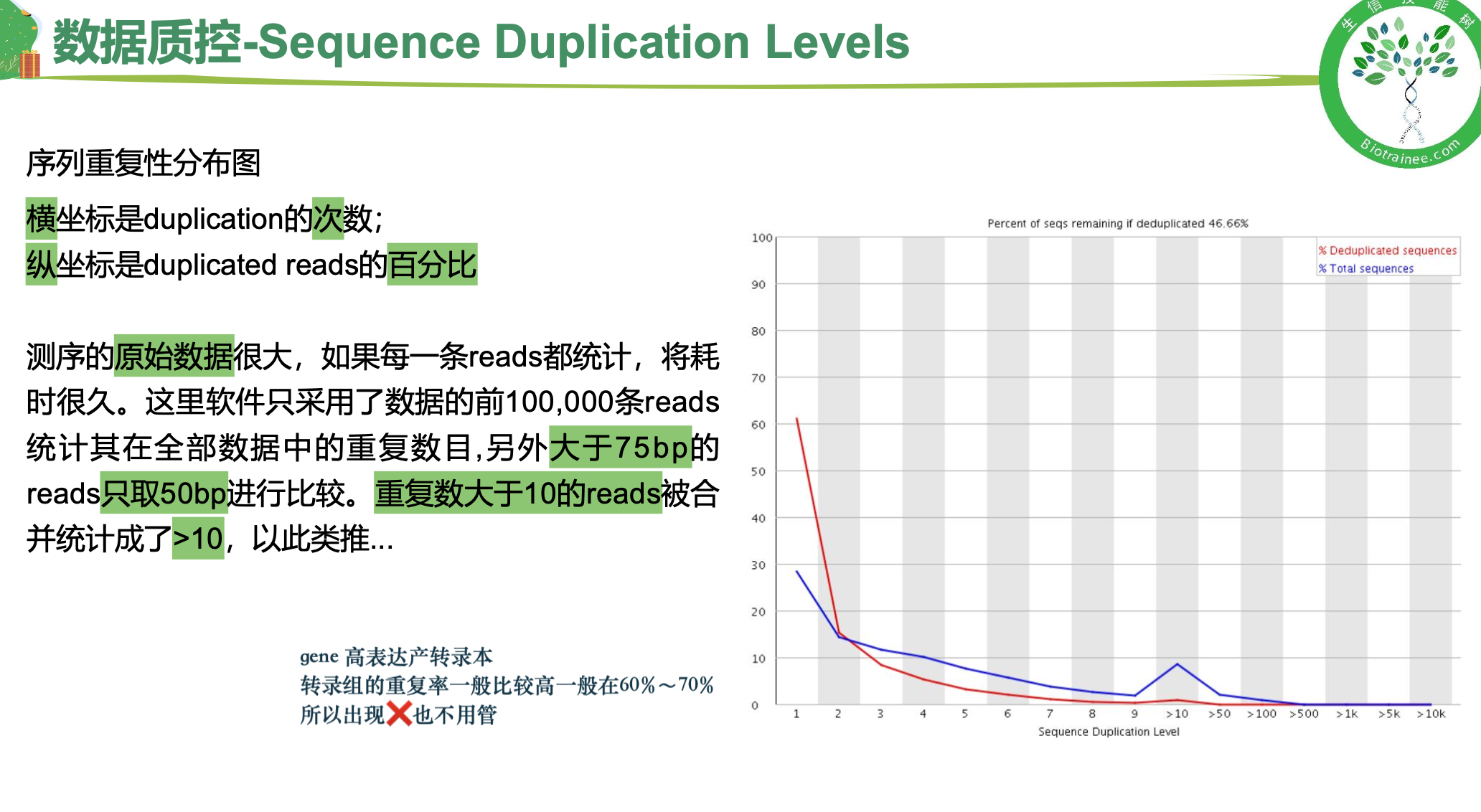
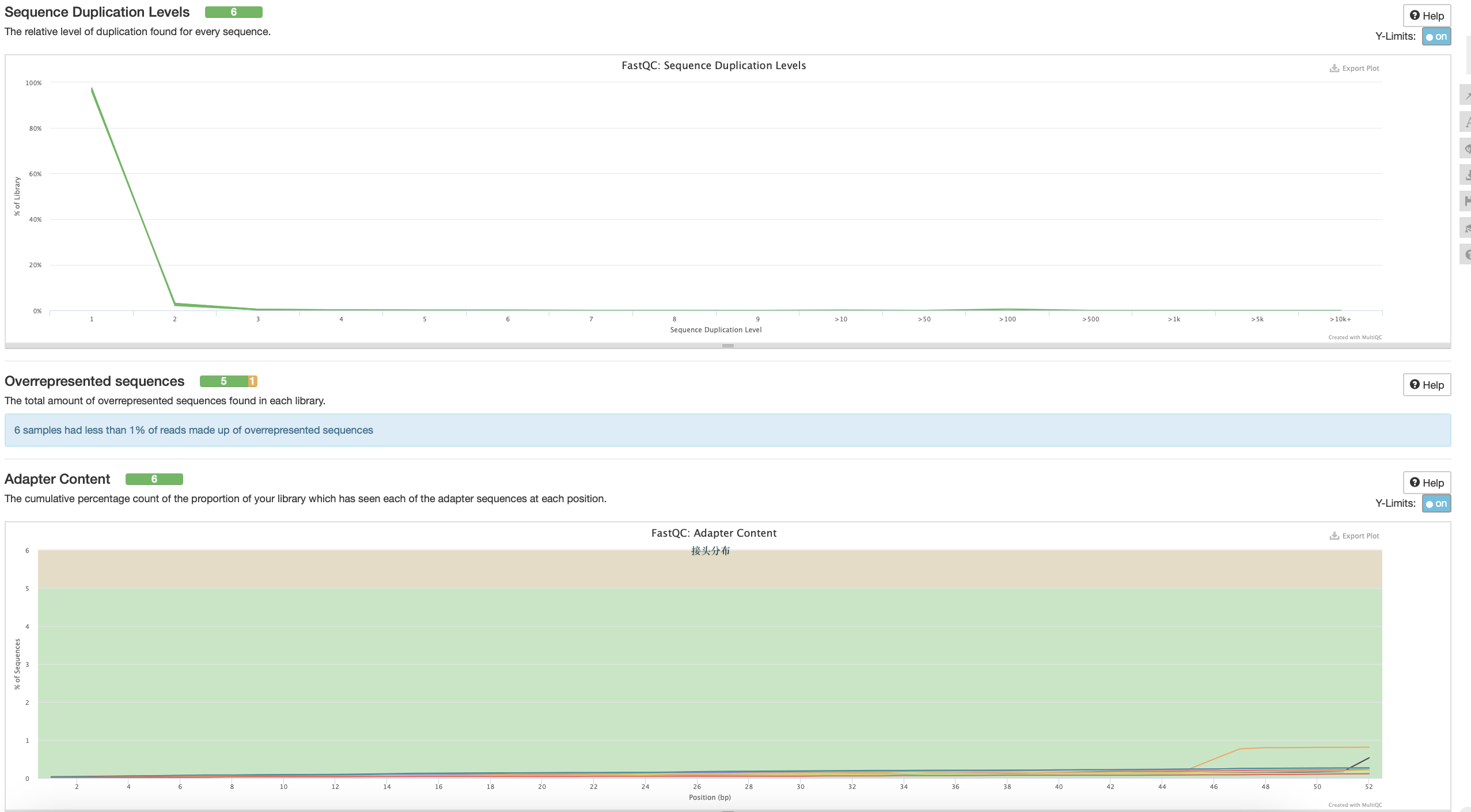
序列重复度
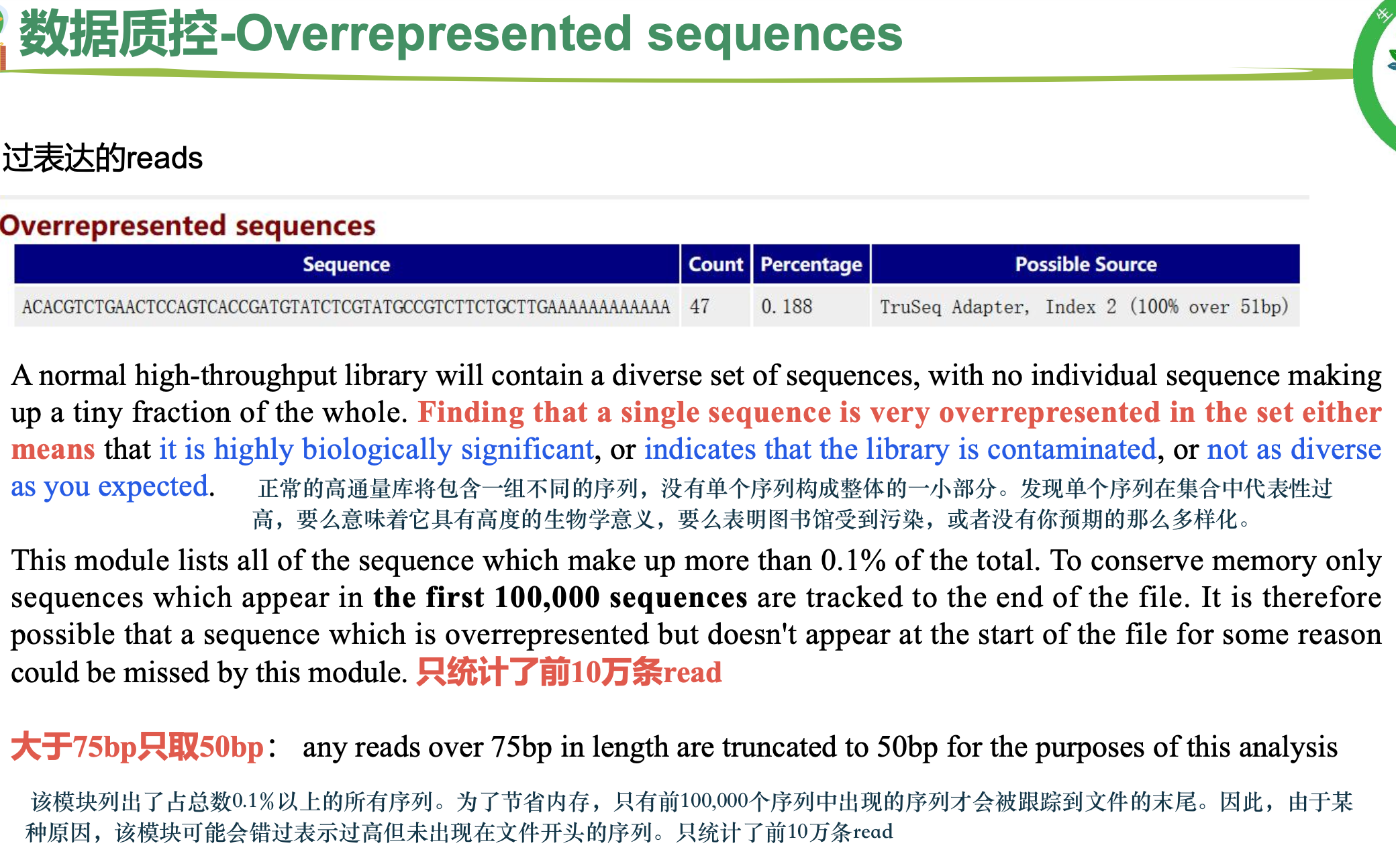
过表达
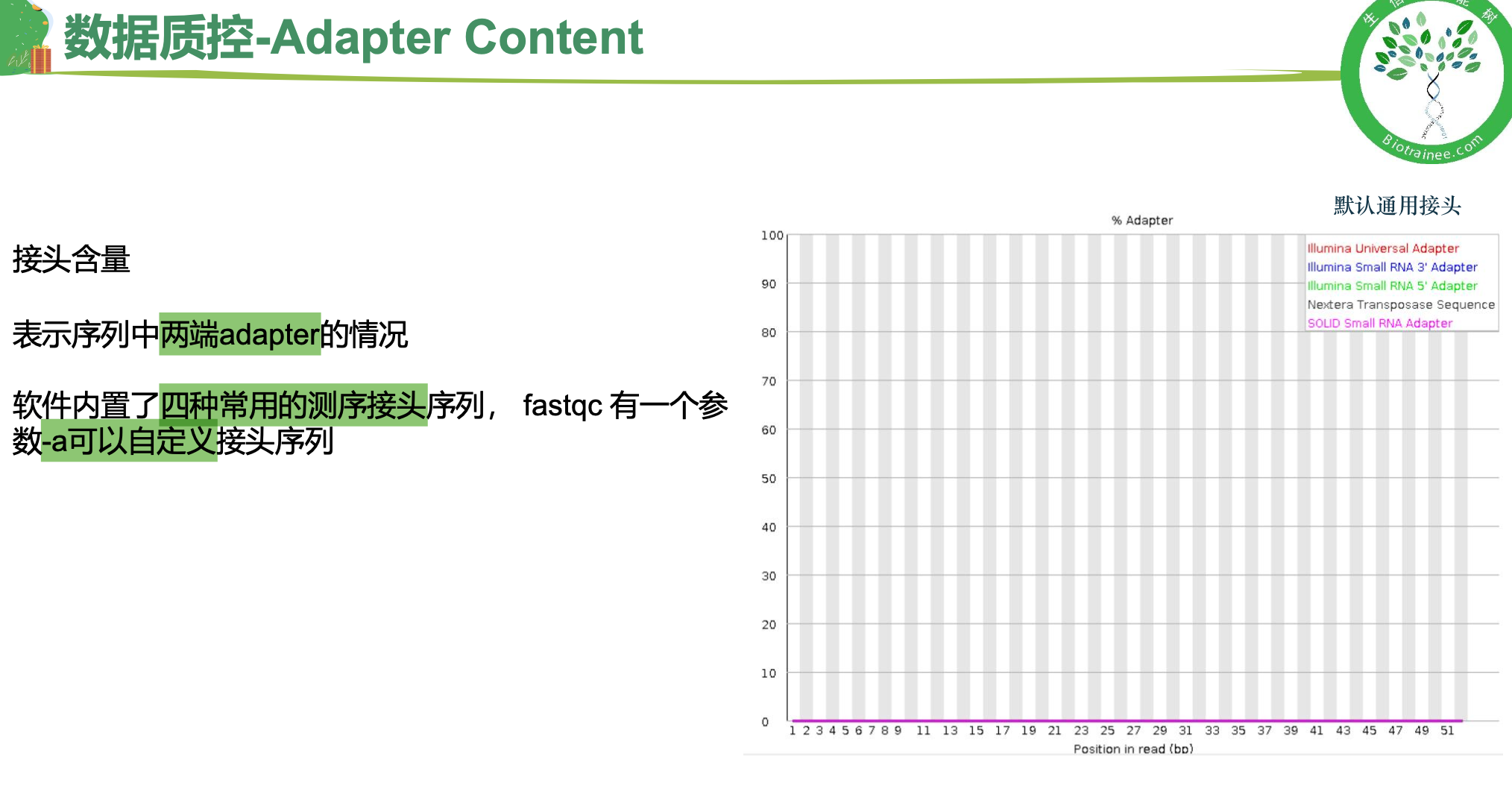
接头含量
一般只看 per base sequence quality 和 per sequence GC content
使用Multi QC整合报告
multiqc *.zip -o ./ #-o 整合到当前目录
再将整合的网页版文件下载到本地 (pic Multi QC)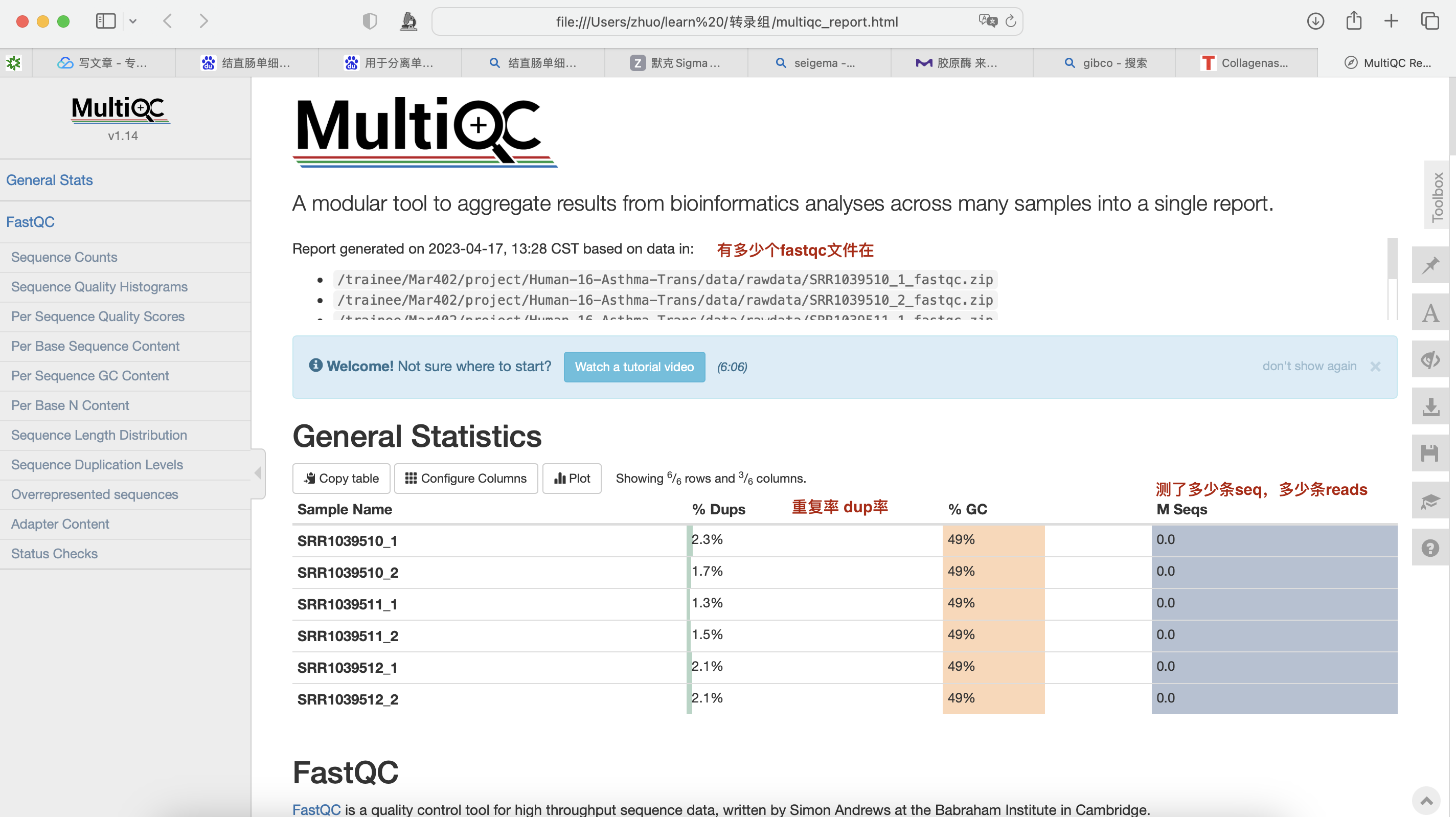
·对于转录组数据中的%Dups只要不超过80%即可
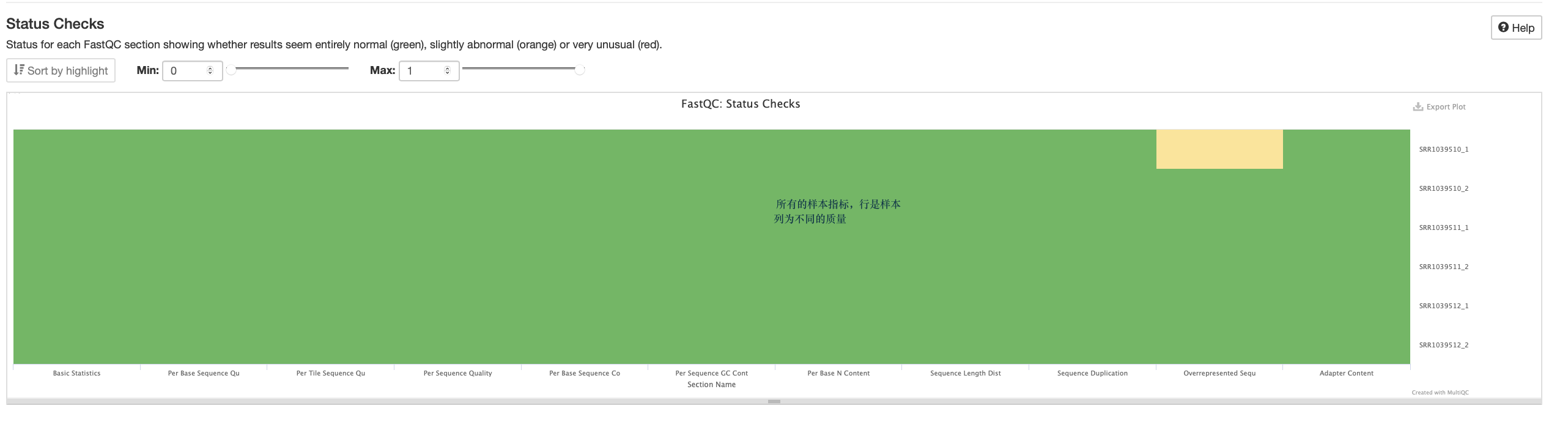
过滤低质量
是否需要过率低质量主要看--per base N content、sequence quality Histograms 、adapter content

过滤低质量

数据过滤常用参数
单个样本过滤低质量运行
(rna) Mar402 20:59:04 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ ../../rawdata/SRR1039510_1.fastq.gz ../../rawdata/SRR1039510_2.fastq.gz
·加载过程···
·加载过程···
·加载过程···
·加载过程···
(rna) Mar402 21:00:23 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls
SRR1039510_1.fastq.gz_trimming_report.txt SRR1039510_2.fastq.gz_trimming_report.txt
SRR1039510_1_val_1_fastqc.html SRR1039510_2_val_2_fastqc.html
SRR1039510_1_val_1_fastqc.zip SRR1039510_2_val_2_fastqc.zip
SRR1039510_1_val_1.fq.gz SRR1039510_2_val_2.fq.gz多个样本过滤低质量运行
# 复杂解析
$ ls /trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/ *_1.fastq.gz #找出所有序列的名字,为了抓取SRR1039510这一段
/trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/SRR1039510_1.fastq.gz
/trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/SRR1039511_1.fastq.gz
/trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/SRR1039512_1.fastq.gz
(rna) Mar402 21:10:21 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls /trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/ *_1.fastq.gz |awk -F'/' '{print $8}' #输出第八个向量,很麻烦需要数,可用下面的方法
SRR1039510_1.fastq.gz
SRR1039511_1.fastq.gz
SRR1039512_1.fastq.gz
(rna) Mar402 21:10:21 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls /trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/ *_1.fastq.gz |awk -F'/' '{print $NF}' #输出最后一个向量
SRR1039510_1.fastq.gz
SRR1039511_1.fastq.gz
SRR1039512_1.fastq.gz
(rna) Mar402 21:10:47 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls /trainee/Mar402/project/Human-16-Asthma-Trans/data/rawdata/ *_1.fastq.gz |awk -F'/' '{print $NF}' |cut -d'_' -f 1 #在此基础上以“-”为分隔符,抓取第一列
SRR1039510
SRR1039511
SRR1039512
(rna) Mar402 21:11:27 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls $HOME/project/Human-16-Asthma-Trans/data/rawdata/ *_1.fastq.gz | awk -F'/' '{print $NF}' | cut -d'_' -f1 >ID
(rna) Mar402 21:17:26 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore #将抓取的序列名称输出为ID
$ cat ID
SRR1039510
SRR1039511
SRR1039512
# 多个样本 vim trim_galore.sh,以下为sh的内容
rawdata=$HOME/project/Human-16-Asthma-Trans/data/rawdata
cleandata=$HOME/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
cat ID | while read id
do
trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ${cleandata} ${rawdata}/${id}_1.fastq.gz ${rawdata}/${id}_2.fastq.gz
done
(rna) Mar402 21:17:38 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ vim trim.sh #将上面的代码写到脚本中
(rna) Mar402 21:22:28 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ less trim.sh #查看脚本中写的代码
(rna) Mar402 21:22:54 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ nohup sh trim.sh >trim.log & #后台运行脚本
[1] 17423
nohup: ignoring input and redirecting stderr to stdout #查看运行 pic123
(rna) Mar402 21:23:29 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore #已经显示Done(已经完成)
$ htop
[1]+ Done nohup sh trim.sh > trim.log
(rna) Mar402 21:24:53 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls #查看
ID SRR1039511_2_val_2_fastqc.html
SRR1039510_1.fastq.gz_trimming_report.txt SRR1039511_2_val_2_fastqc.zip
SRR1039510_1_val_1_fastqc.html SRR1039511_2_val_2.fq.gz
SRR1039510_1_val_1_fastqc.zip SRR1039512_1.fastq.gz_trimming_report.txt
SRR1039510_1_val_1.fq.gz SRR1039512_1_val_1_fastqc.html
SRR1039510_2.fastq.gz_trimming_report.txt SRR1039512_1_val_1_fastqc.zip
SRR1039510_2_val_2_fastqc.html SRR1039512_1_val_1.fq.gz
SRR1039510_2_val_2_fastqc.zip SRR1039512_2.fastq.gz_trimming_report.txt
SRR1039510_2_val_2.fq.gz SRR1039512_2_val_2_fastqc.html
SRR1039511_1.fastq.gz_trimming_report.txt SRR1039512_2_val_2_fastqc.zip
SRR1039511_1_val_1_fastqc.html SRR1039512_2_val_2.fq.gz
SRR1039511_1_val_1_fastqc.zip trim.log
SRR1039511_1_val_1.fq.gz trim.sh
SRR1039511_2.fastq.gz_trimming_report.txt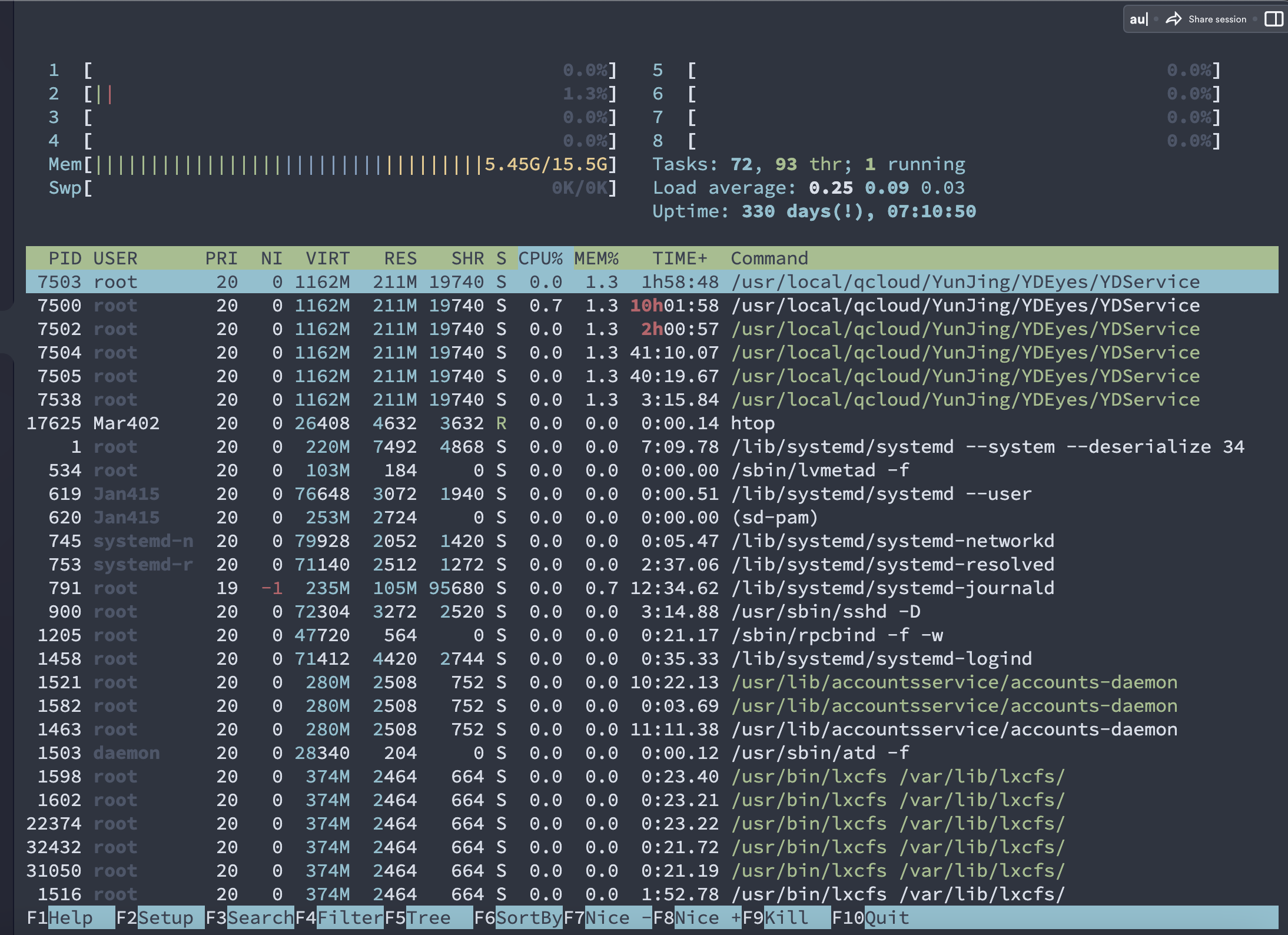
pic123
运行过程中的任务管理
pic1234
#例子
(rna) Mar402 21:25:00 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz
Multicore support not enabled. Proceeding with single-core trimming.
Path to Cutadapt set as: 'cutadapt' (default)
Cutadapt seems to be working fine (tested command 'cutadapt --version')
Cutadapt version: 4.3
single-core operation.
igzip command line interface 2.30.0
igzip detected. Using igzip for decompressing
No quality encoding type selected. Assuming that the data provided uses Sanger encoded Phred scores (default)
Output will be written into the directory: /trainee/Mar402/project/Human-16-Asthma-Trans/data/cleandata/trim_galore/
AUTO-DETECTING ADAPTER TYPE
===========================
Attempting to auto-detect adapter type from the first 1 million sequences of the first file (>> /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz <<)
Found perfect matches for the following adapter sequences:
Adapter type Count Sequence Sequences analysed Percentage
Illumina 1617 AGATCGGAAGAGC 1000000 0.16
Nextera 3 CTGTCTCTTATA 1000000 0.00
smallRNA 1 TGGAATTCTCGG 1000000 0.00
Using Illumina adapter for trimming (count: 1617). Second best hit was Nextera (count: 3)
Writing report to '/trainee/Mar402/project/Human-16-Asthma-Trans/data/cleandata/trim_galore/SRR1039510_1.fastq.gz_trimming_report.txt'
SUMMARISING RUN PARAMETERS
==========================
Input filename: /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz
Trimming mode: paired-end
Trim Galore version: 0.6.10
Cutadapt version: 4.3
Number of cores used for trimming: 1
Quality Phred score cutoff: 20
Quality encoding type selected: ASCII+33
Adapter sequence: 'AGATCGGAAGAGC' (Illumina TruSeq, Sanger iPCR; auto-detected)
Maximum trimming error rate: 0.1 (default)
Maximum number of tolerated Ns: 3
Minimum required adapter overlap (stringency): 3 bp
Minimum required sequence length for both reads before a sequence pair gets removed: 20 bp
Running FastQC on the data once trimming has completed
Output file(s) will be GZIP compressed
Cutadapt seems to be fairly up-to-date (version 4.3). Setting -j 1
Writing final adapter and quality trimmed output to SRR1039510_1_trimmed.fq.gz
>>> Now performing quality (cutoff '-q 20') and adapter trimming in a single pass for the adapter sequence: 'AGATCGGAAGAGC' from file /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz <<<
^Z #输入control z 暂停任务 显示 [1] stop
[1]+ Stopped trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz
(rna) Mar402 21:35:30 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ jobs #jobs 查看
[1]+ Stopped trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz
(rna) Mar402 21:35:47 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ bg %1 #bg (back ground 转向后台运行 +%+项目号(1) )
[1]+ trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz &
(rna) Mar402 21:35:57 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ jobs #jobs 查看 显示running 运行
[1]+ Running trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz &
(rna) Mar402 21:36:08 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ls
ID SRR1039511_2.fastq.gz_trimming_report.txt
SRR1039510_1.fastq.gz_trimming_report.txt SRR1039511_2_val_2_fastqc.html
SRR1039510_1_trimmed.fq.gz SRR1039511_2_val_2_fastqc.zip
SRR1039510_1_val_1_fastqc.html SRR1039511_2_val_2.fq.gz
SRR1039510_1_val_1_fastqc.zip SRR1039512_1.fastq.gz_trimming_report.txt
SRR1039510_1_val_1.fq.gz SRR1039512_1_val_1_fastqc.html
SRR1039510_2.fastq.gz_trimming_report.txt SRR1039512_1_val_1_fastqc.zip
SRR1039510_2_val_2_fastqc.html SRR1039512_1_val_1.fq.gz
SRR1039510_2_val_2_fastqc.zip SRR1039512_2.fastq.gz_trimming_report.txt
SRR1039510_2_val_2.fq.gz SRR1039512_2_val_2_fastqc.html
SRR1039511_1.fastq.gz_trimming_report.txt SRR1039512_2_val_2_fastqc.zip
SRR1039511_1_val_1_fastqc.html SRR1039512_2_val_2.fq.gz
SRR1039511_1_val_1_fastqc.zip trim.log
SRR1039511_1_val_1.fq.gz trim.sh
(rna) Mar402 21:41:27 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ fg %1 #后台转前台 Ctrl+cc 关闭任务或者kill
trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz
^C
(rna) Mar402 21:41:48 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ jobs
#用kill -9 %1 (1为项目号[]中的)杀掉任务
(rna) Mar402 21:45:44 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ jobs
[1]+ Running trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz &
(rna) Mar402 21:45:58 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ kill -9 %1 #杀掉任务
(rna) Mar402 21:46:09 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ jobs #查看
[1]+ Killed trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz
#也可查找编号 杀掉任务
(rna) Mar402 21:49:44 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ps fx #查看代码运行的编号
PID TTY STAT TIME COMMAND
4166 ? S 0:00 sshd: Mar402@pts/2
4167 pts/2 Ss 0:00 \_ -bash
28826 pts/2 T 0:01 \_ perl /trainee/Mar402/miniconda3/envs/rna/bin/trim_galore -q 20 --length 20 --
28833 pts/2 T 0:00 | \_ igzip -d -c /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz
28905 pts/2 R+ 0:00 \_ ps fx
4010 ? Ss 0:00 /lib/systemd/systemd --user
4011 ? S 0:00 \_ (sd-pam)
(rna) Mar402 21:49:52 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ kill -9 28826 #杀掉任务
[1]+ Killed trim_galore -q 20 --length 20 --max_n 3 --stringency 3 --fastqc --paired -o ./ /home/t_rna/data/airway/fastq_raw/SRR1039510_1.fastq.gz /home/t_rna/data/airway/fastq_raw/SRR1039510_2.fastq.gz
(rna) Mar402 21:50:16 ~/project/Human-16-Asthma-Trans/data/cleandata/trim_galore
$ ps fx #任务已经被杀掉
PID TTY STAT TIME COMMAND
4166 ? S 0:00 sshd: Mar402@pts/2
4167 pts/2 Ss 0:00 \_ -bash
29841 pts/2 R+ 0:00 \_ ps fx
4010 ? Ss 0:00 /lib/systemd/systemd --user
4011 ? S 0:00 \_ (sd-pam)
(rna) Mar402 21:52:07 ~/project/Human-16
1234

数据过滤结果
另一个过滤软件fastp 特点快

fastp常用参数
代码对应图 pic345
cd $HOME/project/Human-16-Asthma-Trans/data/cleandata/fastp
(rna) Mar402 21:03:47 ~/project/Human-16-Asthma-Trans/data/cleandata/fastp
$ vim fastp.sh #代码写入脚本
(rna) Mar402 21:11:08 ~/project/Human-16-Asthma-Trans/data/cleandata/fastp
$ nohup sh fastp.sh >fastp.log &
[1] 18011 # 写入日志
nohup: ignoring input and redirecting stderr to stdout
(rna) Mar402 21:11:56 ~/project/Human-16-Asthma-Trans/data/cleandata/fastp
$
[1]+ Done nohup sh fastp.sh > fastp.log #完成
# 定义文件夹:vim fastp.sh
cleandata=$HOME/project/Human-16-Asthma-Trans/data/cleandata/fastp/
rawdata=$HOME/project/Human-16-Asthma-Trans/data/rawdata/
cat ../trim_galore/ID | while read id # 循环
do
fastp -l 20 -q 20 --compression=6 \ #\ 可以换行继续输入命令
-i ${rawdata}/${id}_1.fastq.gz \
-I ${rawdata}/${id}_2.fastq.gz \
-o ${cleandata}/${id}_clean_1.fq.gz \ #输出的文件名称(过滤后reads1)
-O ${cleandata}/${id}_clean_2.fq.gz \
-R ${cleandata}/${id} \ #文件的前缀
-h ${cleandata}/${id}.fastp.html \
-j ${cleandata}/${id}.fastp.json
done
# 运行fastp脚本
nohup sh fastp.sh >fastp.log &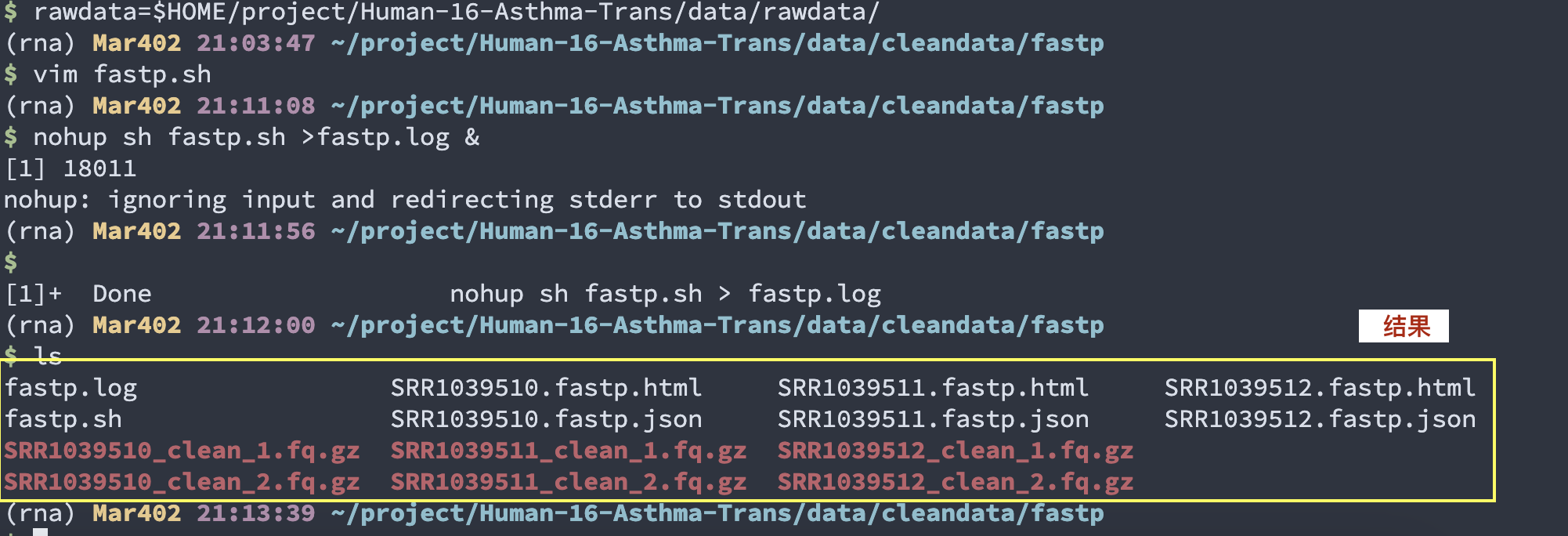
pic345
下载质检报告进行查看
----来自生信技能树----
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
作者已关闭评论
推荐阅读
目录