TCGA分析-数据整理-1
原创title: "三大R包差异分析"
output: html_document
editor_options:
chunk_output_type: console
学习自生信技能树
1.三大R包差异分析
rm(list = ls())
load("GSE218606.Rdata")
proj = "GSE218606"
table(group)
#> group
#> NC OMV2
#> 3 3
#deseq2----
library(DESeq2)
colData <- data.frame(row.names =colnames(exp6),
condition=group)
if(!file.exists(paste0(proj,"_dd.Rdata"))){
dds <- DESeqDataSetFromMatrix(
countData = exp6,
colData = colData,
design = ~ condition)
dds <- DESeq(dds)
save(dds,file = paste0(proj,"_dd.Rdata"))
}
load(file = paste0(proj,"_dd.Rdata"))
class(dds)
#> [1] "DESeqDataSet"
#> attr(,"package")
#> [1] "DESeq2"
res <- results(dds, contrast = c("condition",rev(levels(group))))
#constrast
c("condition",rev(levels(group)))
#> [1] "condition" "OMV2" "NC"
class(res)
#> [1] "DESeqResults"
#> attr(,"package")
#> [1] "DESeq2"
DEG1 <- as.data.frame(res)
library(dplyr)
DEG1 <- arrange(DEG1,pvalue)
DEG1 = na.omit(DEG1)
head(DEG1)
#> baseMean log2FoldChange lfcSE stat pvalue padj
#> PLAU 5924.7264 3.087344 0.1083509 28.49393 1.392927e-178 2.327164e-174
#> IFI6 2259.2701 4.018509 0.1520898 26.42195 7.667321e-154 6.404897e-150
#> CXCL8 559.6508 5.078114 0.1931456 26.29164 2.390236e-152 1.331122e-148
#> NPTX1 2759.8540 2.853715 0.1153558 24.73837 4.135677e-135 1.727369e-131
#> TFPI2 4922.2809 2.559564 0.1045438 24.48318 2.231455e-132 7.456184e-129
#> IFITM1 796.9565 4.005687 0.1667545 24.02146 1.659665e-127 4.621337e-124
#添加change列标记基因上调下调
logFC_t = 2
pvalue_t = 0.05
k1 = (DEG1$pvalue < pvalue_t)&(DEG1$log2FoldChange < -logFC_t);table(k1)
#> k1
#> FALSE TRUE
#> 16430 277
k2 = (DEG1$pvalue < pvalue_t)&(DEG1$log2FoldChange > logFC_t);table(k2)
#> k2
#> FALSE TRUE
#> 16553 154
DEG1$change = ifelse(k1,"DOWN",ifelse(k2,"UP","NOT"))
table(DEG1$change)
#>
#> DOWN NOT UP
#> 277 16276 154
head(DEG1)
#> baseMean log2FoldChange lfcSE stat pvalue padj
#> PLAU 5924.7264 3.087344 0.1083509 28.49393 1.392927e-178 2.327164e-174
#> IFI6 2259.2701 4.018509 0.1520898 26.42195 7.667321e-154 6.404897e-150
#> CXCL8 559.6508 5.078114 0.1931456 26.29164 2.390236e-152 1.331122e-148
#> NPTX1 2759.8540 2.853715 0.1153558 24.73837 4.135677e-135 1.727369e-131
#> TFPI2 4922.2809 2.559564 0.1045438 24.48318 2.231455e-132 7.456184e-129
#> IFITM1 796.9565 4.005687 0.1667545 24.02146 1.659665e-127 4.621337e-124
#> change
#> PLAU UP
#> IFI6 UP
#> CXCL8 UP
#> NPTX1 UP
#> TFPI2 UP
#> IFITM1 UP
#edgeR----
library(edgeR)
dge <- DGEList(counts=exp6,group=group)
dge$samples$lib.size <- colSums(dge$counts)
dge <- calcNormFactors(dge)
design <- model.matrix(~group)
dge <- estimateGLMCommonDisp(dge, design)
dge <- estimateGLMTrendedDisp(dge, design)
dge <- estimateGLMTagwiseDisp(dge, design)
fit <- glmFit(dge, design)
fit <- glmLRT(fit)
DEG2=topTags(fit, n=Inf)
class(DEG2)
#> [1] "TopTags"
#> attr(,"package")
#> [1] "edgeR"
DEG2=as.data.frame(DEG2)
head(DEG2)
#> logFC logCPM LR PValue FDR
#> CXCL1 5.983541 4.946406 772.8412 4.335153e-170 8.381150e-166
#> CXCL8 5.044819 5.125406 749.2813 5.750142e-165 5.558375e-161
#> PLAU 3.059113 8.529890 699.6658 3.534989e-154 2.278064e-150
#> IFI6 3.989729 7.138273 677.7850 2.025911e-149 9.791735e-146
#> NPTX1 2.825230 7.428503 631.8232 2.005855e-139 7.755839e-136
#> TFPI2 2.531606 8.263682 555.5129 7.956607e-123 2.563751e-119
k1 = (DEG2$PValue < pvalue_t)&(DEG2$logFC < -logFC_t)
k2 = (DEG2$PValue < pvalue_t)&(DEG2$logFC > logFC_t)
DEG2$change = ifelse(k1,"DOWN",ifelse(k2,"UP","NOT"))
head(DEG2)
#> logFC logCPM LR PValue FDR change
#> CXCL1 5.983541 4.946406 772.8412 4.335153e-170 8.381150e-166 UP
#> CXCL8 5.044819 5.125406 749.2813 5.750142e-165 5.558375e-161 UP
#> PLAU 3.059113 8.529890 699.6658 3.534989e-154 2.278064e-150 UP
#> IFI6 3.989729 7.138273 677.7850 2.025911e-149 9.791735e-146 UP
#> NPTX1 2.825230 7.428503 631.8232 2.005855e-139 7.755839e-136 UP
#> TFPI2 2.531606 8.263682 555.5129 7.956607e-123 2.563751e-119 UP
table(DEG2$change)
#>
#> DOWN NOT UP
#> 466 18653 214
#limma----
library(limma)
dge <- edgeR::DGEList(counts=exp6)
dge <- edgeR::calcNormFactors(dge)
design <- model.matrix(~group)
v <- voom(dge,design, normalize="quantile")
fit <- lmFit(v, design)
fit= eBayes(fit)
DEG3 = topTable(fit, coef=2, n=Inf)
DEG3 = na.omit(DEG3)
k1 = (DEG3$P.Value < pvalue_t)&(DEG3$logFC < -logFC_t)
k2 = (DEG3$P.Value < pvalue_t)&(DEG3$logFC > logFC_t)
DEG3$change = ifelse(k1,"DOWN",ifelse(k2,"UP","NOT"))
table(DEG3$change)
#>
#> DOWN NOT UP
#> 295 18790 248
head(DEG3)
#> logFC AveExpr t P.Value adj.P.Val B change
#> PLAU 3.000591 7.792455 35.81687 2.129933e-12 4.117800e-08 18.76396 UP
#> TFPI2 2.480469 7.746460 32.72240 5.503366e-12 5.319829e-08 17.94908 UP
#> NPTX1 2.861412 6.776749 28.54269 2.305755e-11 1.485905e-07 16.55314 UP
#> IFI6 4.083173 5.961048 26.22506 5.589770e-11 2.161341e-07 15.58656 UP
#> SECTM1 3.683304 4.841097 24.48167 1.145860e-10 3.692153e-07 14.71278 UP
#> FAM167A 2.896689 4.933178 23.98668 1.417644e-10 3.915330e-07 14.63831 UP
tj = data.frame(deseq2 = as.integer(table(DEG1$change)),
edgeR = as.integer(table(DEG2$change)),
limma_voom = as.integer(table(DEG3$change)),
row.names = c("down","not","up")
);tj
#> deseq2 edgeR limma_voom
#> down 277 466 295
#> not 16276 18653 18790
#> up 154 214 248
save(DEG1,DEG2,DEG3,group,tj,file = paste0(proj,"_DEG.Rdata"))2.可视化
library(ggplot2)
library(tinyarray)
exp6[1:4,1:4]
#> NC_1 NC_2 NC_3 OMV2_1
#> FN1 645312 500253 534583 324566
#> EEF1A1 195214 174595 188659 196505
#> MT-CO1 284380 167314 188611 192357
#> ACTB 161056 133202 144670 161598
# cpm 去除文库大小的影响
dat = log2(cpm(exp6)+1)
pca.plot = draw_pca(dat,group);pca.plot
save(pca.plot,file = paste0(proj,"_pcaplot.Rdata"))
cg1 = rownames(DEG1)[DEG1$change !="NOT"]
cg2 = rownames(DEG2)[DEG2$change !="NOT"]
cg3 = rownames(DEG3)[DEG3$change !="NOT"]
h1 = draw_heatmap(dat[cg1,],group)
h2 = draw_heatmap(dat[cg2,],group)
h3 = draw_heatmap(dat[cg3,],group)
v1 = draw_volcano(DEG1,pkg = 1,logFC_cutoff = logFC_t)
v2 = draw_volcano(DEG2,pkg = 2,logFC_cutoff = logFC_t)
v3 = draw_volcano(DEG3,pkg = 3,logFC_cutoff = logFC_t)
library(patchwork)
(h1 + h2 + h3) / (v1 + v2 + v3) +plot_layout(guides = 'collect') &theme(legend.position = "none")
ggsave(paste0(proj,"_heat_vo.png"),width = 15,height = 10)3.三大R包差异基因对比
UP=function(df){
rownames(df)[df$change=="UP"]
}
DOWN=function(df){
rownames(df)[df$change=="DOWN"]
}
up = intersect(intersect(UP(DEG1),UP(DEG2)),UP(DEG3))
down = intersect(intersect(DOWN(DEG1),DOWN(DEG2)),DOWN(DEG3))
dat = log2(cpm(exp6)+1)
dat=as.data.frame(dat)
hp = draw_heatmap(dat[c(up,down),],group)
#上调、下调基因分别画维恩图
up_genes = list(Deseq2 = UP(DEG1),
edgeR = UP(DEG2),
limma = UP(DEG3))
down_genes = list(Deseq2 = DOWN(DEG1),
edgeR = DOWN(DEG2),
limma = DOWN(DEG3))
up.plot <- draw_venn(up_genes,"UPgene")
down.plot <- draw_venn(down_genes,"DOWNgene")
#维恩图拼图,终于搞定
library(patchwork)
#up.plot + down.plot
# 拼图
pca.plot + hp+up.plot +down.plot+ plot_layout(guides = "collect")
ggsave(paste0(proj,"_heat_ve_pca.png"),width = 15,height = 10)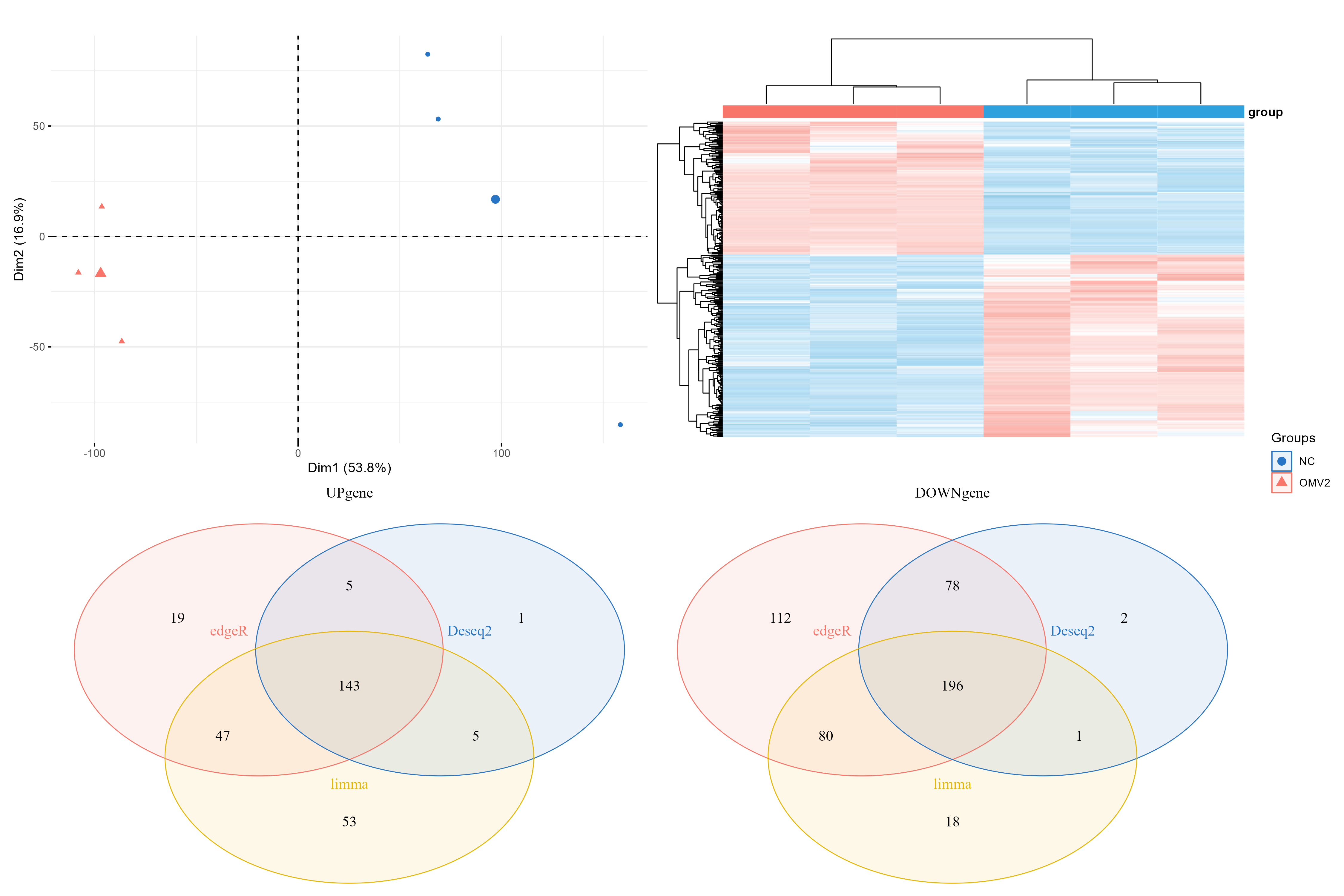
分组聚类的热图
library(ComplexHeatmap)
library(circlize)
col_fun = colorRamp2(c(-2, 0, 2), c("#2fa1dd", "white", "#f87669"))
top_annotation = HeatmapAnnotation(
cluster = anno_block(gp = gpar(fill = c("#f87669","#2fa1dd")),
labels = levels(group),
labels_gp = gpar(col = "white", fontsize = 12)))
m = Heatmap(t(scale(t(exp6[c(up,down),]))),name = " ",
col = col_fun,
top_annotation = top_annotation,
column_split = group,
show_heatmap_legend = T,
border = F,
show_column_names = F,
show_row_names = F,
use_raster = F,
cluster_column_slices = F,
column_title = NULL)
m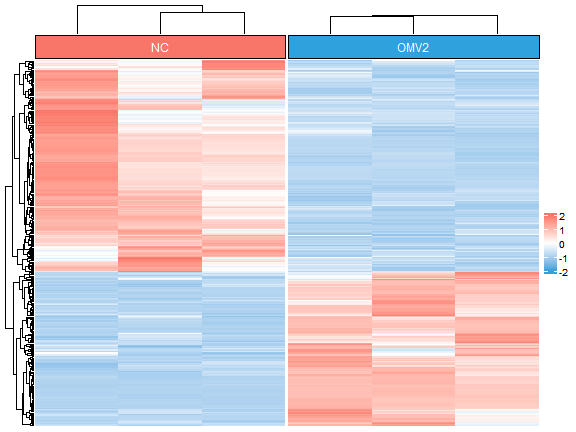
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录
